Difference between revisions of "Zur"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of zinc homeostasis<br/>(''[[yciC]], [[znuA]]-[[znuC]]-[[znuB]]'') | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of zinc homeostasis<br/>(''[[yciC]], [[znuA]]-[[znuC]]-[[znuB]]'') | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU25100 zur] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Regulatory function of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Regulatory function of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress]''' | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqfW]]'', ''[[yqfU]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqfW]]'', ''[[yqfU]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU25100 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU25100 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU25100 Advanced_DNA] |
|- | |- | ||
|- | |- | ||
Revision as of 13:19, 13 May 2013
- Description: transcriptional repressor, regulates zinc homeostasis
| Gene name | zur |
| Synonyms | yqfV |
| Essential | no |
| Product | transcriptional repressor (Fur family) |
| Function | regulation of zinc homeostasis (yciC, znuA-znuC-znuB) |
| Gene expression levels in SubtiExpress: zur | |
| Regulatory function of this protein in SubtiPathways: Stress | |
| MW, pI | 16 kDa, 4.856 |
| Gene length, protein length | 435 bp, 145 aa |
| Immediate neighbours | yqfW, yqfU |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 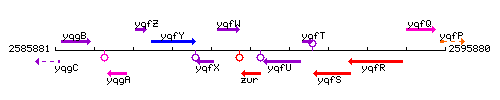
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed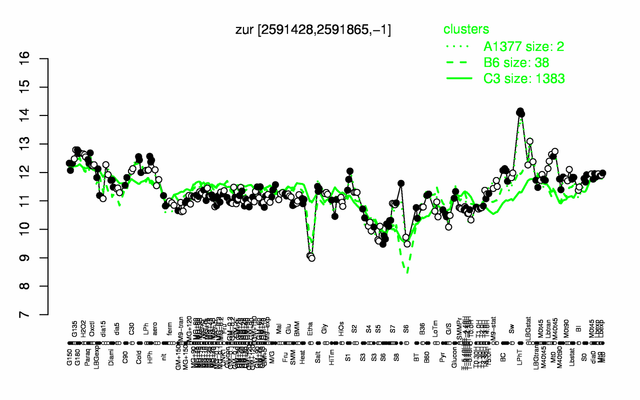
| |
Contents
Categories containing this gene/protein
trace metal homeostasis (Cu, Zn, Ni, Mn, Mo), transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
The Zur regulon
The gene
Basic information
- Locus tag: BSU25100
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Fur family
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on ser/ thr/ tyr PubMed
- Cofactor(s):
- Effectors of protein activity:
- sequential binding of zinc (two atoms per monomer) results in the activation of Zur and thus in repression of the genes of the Zur regulon PubMed
- Interactions:
- forms dimers PubMed
Database entries
- Structure:
- UniProt: P54479
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Zhen Ma, Scott E Gabriel, John D Helmann
Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur.
Nucleic Acids Res: 2011, 39(21);9130-8
[PubMed:21821657]
[WorldCat.org]
[DOI]
(I p)
Scott E Gabriel, John D Helmann
Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions.
J Bacteriol: 2009, 191(19);6116-22
[PubMed:19648245]
[WorldCat.org]
[DOI]
(I p)
Scott E Gabriel, Faith Miyagi, Ahmed Gaballa, John D Helmann
Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur.
J Bacteriol: 2008, 190(10);3482-8
[PubMed:18344368]
[WorldCat.org]
[DOI]
(I p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Mayuree Fuangthong, John D Helmann
Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis.
J Bacteriol: 2003, 185(21);6348-57
[PubMed:14563870]
[WorldCat.org]
[DOI]
(P p)
Ekaterina M Panina, Andrey A Mironov, Mikhail S Gelfand
Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins.
Proc Natl Acad Sci U S A: 2003, 100(17);9912-7
[PubMed:12904577]
[WorldCat.org]
[DOI]
(P p)
Ahmed Gaballa, Tao Wang, Rick W Ye, John D Helmann
Functional analysis of the Bacillus subtilis Zur regulon.
J Bacteriol: 2002, 184(23);6508-14
[PubMed:12426338]
[WorldCat.org]
[DOI]
(P p)
A Gaballa, J D Helmann
Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis.
J Bacteriol: 1998, 180(22);5815-21
[PubMed:9811636]
[WorldCat.org]
[DOI]
(P p)