Difference between revisions of "MreBH"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || [[cell shape]] determation | |style="background:#ABCDEF;" align="center"|'''Function''' || [[cell shape]] determation | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU14470 mreBH] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/MreBH MreBH] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/MreBH MreBH] | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ykpC]]'', ''[[abh]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ykpC]]'', ''[[abh]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU14470 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU14470 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU14470 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:mreBH_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:mreBH_context.gif]] | ||
Revision as of 12:43, 13 May 2013
- Description: cell shape-determining protein, forms filaments, the polymers control/restrict the mobility of the cell wall elongation enzyme comple
| Gene name | mreBH |
| Synonyms | |
| Essential | no |
| Product | cell shape-determining protein |
| Function | cell shape determation |
| Gene expression levels in SubtiExpress: mreBH | |
| Interactions involving this protein in SubtInteract: MreBH | |
| MW, pI | 35 kDa, 5.239 |
| Gene length, protein length | 1005 bp, 335 aa |
| Immediate neighbours | ykpC, abh |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 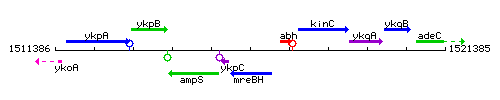
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed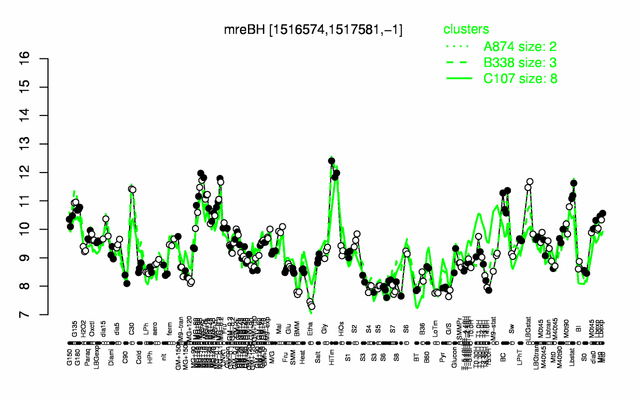
| |
Contents
Categories containing this gene/protein
cell shape, heat shock proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14470
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- forms ring-shaped filaments in a heterologous system PubMed
- Protein family: ftsA/mreB family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- during logarithmic growth, MreBH forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis PubMed
- forms transverse bands as cells enter the stationary phase PubMed
- close to the inner surface of the cytoplasmic membrane PubMed
- reports on helical structures formed by MreBH PubMed seem to be misinterpretation of data PubMed
- normal localization depends on the presence of glucolipids, MreB forms irregular clusters in an ugtP mutant PubMed
Database entries
- Structure:
- UniProt: P39763
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Localization
Satoshi Matsuoka, Minako Chiba, Yu Tanimura, Michihiro Hashimoto, Hiroshi Hara, Kouji Matsumoto
Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids.
Genes Genet Syst: 2011, 86(5);295-304
[PubMed:22362028]
[WorldCat.org]
[DOI]
(I p)
Ethan C Garner, Remi Bernard, Wenqin Wang, Xiaowei Zhuang, David Z Rudner, Tim Mitchison
Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis.
Science: 2011, 333(6039);222-5
[PubMed:21636745]
[WorldCat.org]
[DOI]
(I p)
Julia Domínguez-Escobar, Arnaud Chastanet, Alvaro H Crevenna, Vincent Fromion, Roland Wedlich-Söldner, Rut Carballido-López
Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria.
Science: 2011, 333(6039);225-8
[PubMed:21636744]
[WorldCat.org]
[DOI]
(I p)
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
Hervé Joël Defeu Soufo, Peter L Graumann
Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB.
Mol Microbiol: 2006, 62(5);1340-56
[PubMed:17064365]
[WorldCat.org]
[DOI]
(P p)
Other original publications