Difference between revisions of "SigH"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || [[transcription]] of early stationary phase genes <br/>(sporulation, competence) | |style="background:#ABCDEF;" align="center"|'''Function''' || [[transcription]] of early stationary phase genes <br/>(sporulation, competence) | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU00980 sigH] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SigH SigH] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SigH SigH] | ||
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yacP]]'', ''[[rpmGB]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yacP]]'', ''[[rpmGB]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU00980 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU00980 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU00980 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:sigH_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:sigH_context.gif]] | ||
Revision as of 11:57, 13 May 2013
- Description: RNA polymerase sigma factor SigH
| Gene name | sigH |
| Synonyms | spo0H |
| Essential | no |
| Product | RNA polymerase sigma factor SigH |
| Function | transcription of early stationary phase genes (sporulation, competence) |
| Gene expression levels in SubtiExpress: sigH | |
| Interactions involving this protein in SubtInteract: SigH | |
| MW, pI | 25 kDa, 5.146 |
| Gene length, protein length | 654 bp, 218 aa |
| Immediate neighbours | yacP, rpmGB |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 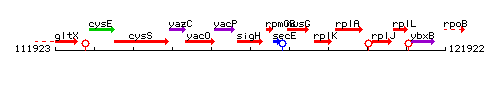
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control
This gene is a member of the following regulons
The SigH regulon
The gene
Basic information
- Locus tag: BSU00980
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P17869
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: sigH (DBTBS)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
The SigH regulon
Original Publications
Imke G de Jong, Jan-Willem Veening, Oscar P Kuipers
Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation.
J Bacteriol: 2010, 192(8);2053-67
[PubMed:20154131]
[WorldCat.org]
[DOI]
(I p)
C Eymann, M Hecker
Induction of sigma(B)-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis.
FEMS Microbiol Lett: 2001, 199(2);221-7
[PubMed:11377871]
[WorldCat.org]
[DOI]
(P p)
C M Buckner, C P Moran
A region in Bacillus subtilis sigmaH required for Spo0A-dependent promoter activity.
J Bacteriol: 1998, 180(18);4987-90
[PubMed:9733708]
[WorldCat.org]
[DOI]
(P p)
P A Levin, R Losick
Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis.
Genes Dev: 1996, 10(4);478-88
[PubMed:8600030]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promote.
J Bacteriol: 1995, 177(23);6999-7002
[PubMed:7592498]
[WorldCat.org]
[DOI]
(P p)
T Bird, D Burbulys, J J Wu, M A Strauch, J A Hoch, G B Spiegelman
The effect of supercoiling on the in vitro transcription of the spoIIA operon from Bacillus subtilis.
Biochimie: 1992, 74(7-8);627-34
[PubMed:1391042]
[WorldCat.org]
[DOI]
(P p)
J Healy, J Weir, I Smith, R Losick
Post-transcriptional control of a sporulation regulatory gene encoding transcription factor sigma H in Bacillus subtilis.
Mol Microbiol: 1991, 5(2);477-87
[PubMed:1904128]
[WorldCat.org]
[DOI]
(P p)
J Weir, M Predich, E Dubnau, G Nair, I Smith
Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor.
J Bacteriol: 1991, 173(2);521-9
[PubMed:1898930]
[WorldCat.org]
[DOI]
(P p)
D Daniels, P Zuber, R Losick
Two amino acids in an RNA polymerase sigma factor involved in the recognition of adjacent base pairs in the -10 region of a cognate promoter.
Proc Natl Acad Sci U S A: 1990, 87(20);8075-9
[PubMed:2122453]
[WorldCat.org]
[DOI]
(P p)
U Bai, M Lewandoski, E Dubnau, I Smith
Temporal regulation of the Bacillus subtilis early sporulation gene spo0F.
J Bacteriol: 1990, 172(9);5432-9
[PubMed:2118512]
[WorldCat.org]
[DOI]
(P p)
K M Tatti, H L Carter, A Moir, C P Moran
Sigma H-directed transcription of citG in Bacillus subtilis.
J Bacteriol: 1989, 171(11);5928-32
[PubMed:2509422]
[WorldCat.org]
[DOI]
(P p)
K J Jaacks, J Healy, R Losick, A D Grossman
Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis.
J Bacteriol: 1989, 171(8);4121-9
[PubMed:2502532]
[WorldCat.org]
[DOI]
(P p)
P Zuber, J Healy, H L Carter, S Cutting, C P Moran, R Losick
Mutation changing the specificity of an RNA polymerase sigma factor.
J Mol Biol: 1989, 206(4);605-14
[PubMed:2500529]
[WorldCat.org]
[DOI]
(P p)
H L Carter, L F Wang, R H Doi, C P Moran
rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis.
J Bacteriol: 1988, 170(4);1617-21
[PubMed:3127379]
[WorldCat.org]
[DOI]
(P p)
C Ray, M Igo, W Shafer, R Losick, C P Moran
Suppression of ctc promoter mutations in Bacillus subtilis.
J Bacteriol: 1988, 170(2);900-7
[PubMed:3123466]
[WorldCat.org]
[DOI]
(P p)
P Zuber, R Losick
spoOH: a developmental regulatory gene for promoter utilization in Bacillus subtilis.
Cold Spring Harb Symp Quant Biol: 1985, 50;483-8
[PubMed:3007003]
[WorldCat.org]
[DOI]
(P p)
W C Johnson, C P Moran, R Losick
Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene.
Nature: 1983, 302(5911);800-4
[PubMed:6405278]
[WorldCat.org]
[DOI]
(P p)