Difference between revisions of "MinJ"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 90: | Line 86: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| − | ** ([[MinC]]-[[MinD]])-[[MinJ]]-[[DivIVA]] {{PubMed|20352045}} | + | ** ([[MinC]]-[[MinD]])-[[MinJ]]-[[DivIVA]] {{PubMed|20352045}}, [[MinJ]] binds to the N-terminal lipid-binding domain of [[DivIVA]] {{PubMed|23264578}} |
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** forms rings at the division septum {{PubMed|22108385}} | ** forms rings at the division septum {{PubMed|22108385}} | ||
| − | ** cell membrane | + | ** cell membrane {{PubMed|23264578}} |
=== Database entries === | === Database entries === | ||
| Line 146: | Line 142: | ||
<pubmed> 19884039 </pubmed> | <pubmed> 19884039 </pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>19019154, 16030230,18976281, 20352045, 18567663 22108385 22457634</pubmed> | + | <pubmed>19019154, 16030230,18976281, 20352045, 18567663 22108385 22457634 23264578</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:21, 3 January 2013
- Description: topological determinant of cell division, part of the Min system (with DivIVA, MinC, MinD), Noc and the Min system ensure the efficient utilization of the division site at midcell in by ensuring Z ring placement
| Gene name | minJ |
| Synonyms | yvjD, swrAB |
| Essential | no |
| Product | topological determinant of cell division |
| Function | cell division site selection |
| Gene expression levels in SubtiExpress: minJ | |
| Interactions involving this protein in SubtInteract: MinJ | |
| MW, pI | 43 kDa, 9.057 |
| Gene length, protein length | 1191 bp, 397 aa |
| Immediate neighbours | yvkA, swrAA/1 |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed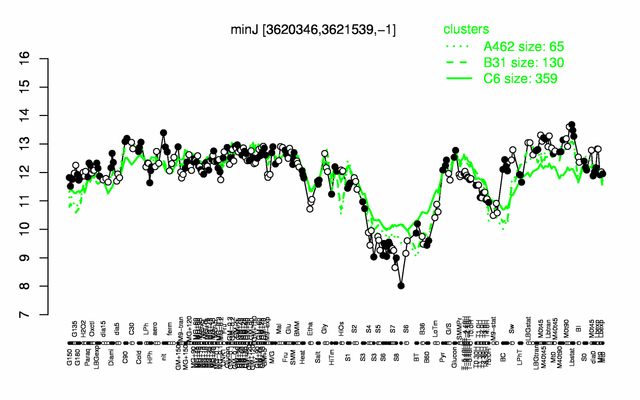
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35220
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: The Min system prevents minicell formation adjacent to recently completed division sites by promoting the disassembly of the cytokinetic ring, thereby ensuring that cell division occurs only once per cell cycle PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O34375
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Daniel Kearns, Indiana University, Bloomington, USA, homepage
Marc Bramkamp, University of Cologne, Cologne, Germany homepage
Your additional remarks
References
Reviews
Original Publications
Suey van Baarle, Ilkay Nazli Celik, Karan Gautam Kaval, Marc Bramkamp, Leendert W Hamoen, Sven Halbedel
Protein-protein interaction domains of Bacillus subtilis DivIVA.
J Bacteriol: 2013, 195(5);1012-21
[PubMed:23264578]
[WorldCat.org]
[DOI]
(I p)
Christopher D A Rodrigues, Elizabeth J Harry
The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization.
PLoS Genet: 2012, 8(3);e1002561
[PubMed:22457634]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Marcella L Erb, James A Gregory, Jared Silverman, Kit Pogliano, Joe Pogliano, Kumaran S Ramamurthi
Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis.
mBio: 2011, 2(6);
[PubMed:22108385]
[WorldCat.org]
[DOI]
(I e)
Suey van Baarle, Marc Bramkamp
The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly.
PLoS One: 2010, 5(3);e9850
[PubMed:20352045]
[WorldCat.org]
[DOI]
(I e)
Marc Bramkamp, Robyn Emmins, Louise Weston, Catriona Donovan, Richard A Daniel, Jeff Errington
A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD.
Mol Microbiol: 2008, 70(6);1556-69
[PubMed:19019154]
[WorldCat.org]
[DOI]
(I p)
Joyce E Patrick, Daniel B Kearns
MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis.
Mol Microbiol: 2008, 70(5);1166-79
[PubMed:18976281]
[WorldCat.org]
[DOI]
(I p)
Cinzia Calvio, Cecilia Osera, Giuseppe Amati, Alessandro Galizzi
Autoregulation of swrAA and motility in Bacillus subtilis.
J Bacteriol: 2008, 190(16);5720-8
[PubMed:18567663]
[WorldCat.org]
[DOI]
(I p)
Cinzia Calvio, Francesco Celandroni, Emilia Ghelardi, Giuseppe Amati, Sara Salvetti, Fabrizio Ceciliani, Alessandro Galizzi, Sonia Senesi
Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon.
J Bacteriol: 2005, 187(15);5356-66
[PubMed:16030230]
[WorldCat.org]
[DOI]
(P p)