Difference between revisions of "FlhG"
(No difference)
| |
Revision as of 17:39, 30 November 2012
- Description: GTPase activating protein, activates FlhF
| Gene name | ylxH |
| Synonyms | |
| Essential | no |
| Product | GTPase activating protein |
| Function | activation of FlhF |
| Gene expression levels in SubtiExpress: ylxH | |
| Interactions involving this protein in SubtInteract: YlxH | |
| MW, pI | 33 kDa, 9.648 |
| Gene length, protein length | 894 bp, 298 aa |
| Immediate neighbours | flhF, cheB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 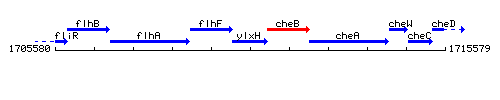
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, resistance against toxins/ antibiotics
This gene is a member of the following regulons
CodY regulon, SigD regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU16410
Phenotypes of a mutant
- susceptible to acriflavine and ethidium bromide, and severe growth inhibition as surfactin concentration increased up to 100ug/ml.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family: View classification (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (Homogeneous) PubMed
Database entries
- Structure:
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References