Difference between revisions of "EngA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || ribosome assembly | |style="background:#ABCDEF;" align="center"|'''Function''' || ribosome assembly | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU22840 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU22840 engA] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 48 kDa, 5.248 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 48 kDa, 5.248 | ||
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 78: | Line 74: | ||
* '''Protein family:''' EngA subfamily (according to Swiss-Prot) Era/Obg family | * '''Protein family:''' EngA subfamily (according to Swiss-Prot) Era/Obg family | ||
| − | * '''Paralogous protein(s):''' [[Era]], [[Obg | + | * '''Paralogous protein(s):''' [[Era]], [[Obg]], [[YsxC]], [[RbgA]], [[YqeH]], [[ThdF]], [[YyaF]], [[YnbA]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 112: | Line 108: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=engA_2390206_2391516_-1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=engA_2390206_2391516_-1 engA] {{PubMed|22383849}} |
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
| Line 127: | Line 123: | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| − | ** for expression, purification in ''E. coli'' with N-terminal His-tag, in [[pWH844]]: pGP846, available in [[Stülke]] lab | + | ** for expression, purification in ''E. coli'' with N-terminal His-tag, in [[pWH844]]: pGP846, available in [[Jörg Stülke]]'s lab |
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| Line 148: | Line 144: | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>12427945, 16997968,16894162 16682769 | + | <pubmed>12427945, 16997968,16894162 16682769 23056455</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:57, 14 October 2012
- Description: GTPase essential for ribosome 50S subunit assembly
| Gene name | yphC |
| Synonyms | engA |
| Essential | yes PubMed |
| Product | GTPase |
| Function | ribosome assembly |
| Gene expression levels in SubtiExpress: engA | |
| MW, pI | 48 kDa, 5.248 |
| Gene length, protein length | 1308 bp, 436 aa |
| Immediate neighbours | gpsA, seaA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 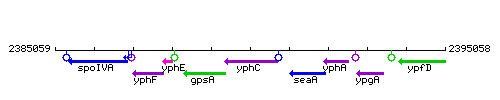
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed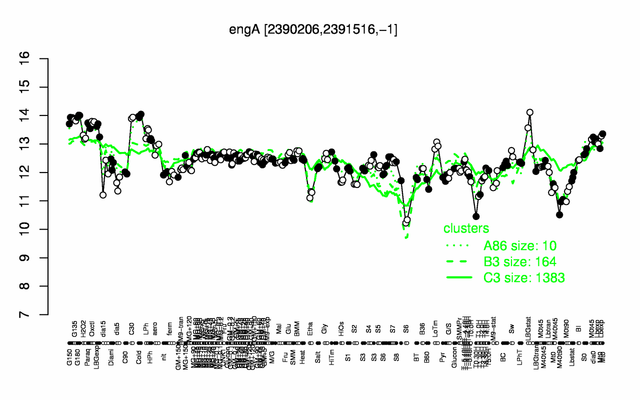
| |
Contents
Categories containing this gene/protein
translation, essential genes, GTP-binding proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22840
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Binds and hydrolyzes GTP and readily exchanges GDP for GTP
- Protein family: EngA subfamily (according to Swiss-Prot) Era/Obg family
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2HJG (complex with GDP)
- UniProt: P50743
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- for expression, purification in E. coli with N-terminal His-tag, in pWH844: pGP846, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Naotake Ogasawara, Nara, Japan
Your additional remarks
References
Reviews
Original publications
Anne-Emmanuelle Foucher, Jean-Baptiste Reiser, Christine Ebel, Dominique Housset, Jean-Michel Jault
Potassium acts as a GTPase-activating element on each nucleotide-binding domain of the essential Bacillus subtilis EngA.
PLoS One: 2012, 7(10);e46795
[PubMed:23056455]
[WorldCat.org]
[DOI]
(I p)
Laura Schaefer, William C Uicker, Catherine Wicker-Planquart, Anne-Emmanuelle Foucher, Jean-Michel Jault, Robert A Britton
Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis.
J Bacteriol: 2006, 188(23);8252-8
[PubMed:16997968]
[WorldCat.org]
[DOI]
(P p)
Stephen P Muench, Ling Xu, Svetlana E Sedelnikova, David W Rice
The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding.
Proc Natl Acad Sci U S A: 2006, 103(33);12359-64
[PubMed:16894162]
[WorldCat.org]
[DOI]
(P p)
Ling Xu, Stephen P Muench, Anna Roujeinikova, Svetlana E Sedelnikova, David W Rice
Cloning, purification and preliminary crystallographic analysis of the Bacillus subtilis GTPase YphC-GDP complex.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2006, 62(Pt 5);435-7
[PubMed:16682769]
[WorldCat.org]
[DOI]
(I p)
Takuya Morimoto, Pek Chin Loh, Tomohiro Hirai, Kei Asai, Kazuo Kobayashi, Shigeki Moriya, Naotake Ogasawara
Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis.
Microbiology (Reading): 2002, 148(Pt 11);3539-3552
[PubMed:12427945]
[WorldCat.org]
[DOI]
(P p)