Difference between revisions of "CydB"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || respiration | |style="background:#ABCDEF;" align="center"|'''Function''' || respiration | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU38750 cydB] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 37 kDa, 9.48 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 37 kDa, 9.48 | ||
Revision as of 17:23, 7 August 2012
- Description: cytochrome bd ubiquinol oxidase (subunit II), high affinity terminal oxidase
| Gene name | cydB |
| Synonyms | yxkL |
| Essential | no |
| Product | cytochrome bd ubiquinol oxidase (subunit II), high affinity terminal oxidase |
| Function | respiration |
| Gene expression levels in SubtiExpress: cydB | |
| MW, pI | 37 kDa, 9.48 |
| Gene length, protein length | 1014 bp, 338 aa |
| Immediate neighbours | cydC, cydA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 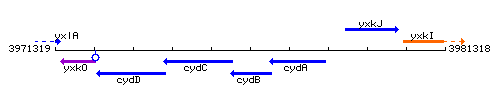
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
respiration, membrane proteins
This gene is a member of the following regulons
CcpA regulon, ResD regulon, Rex regulon
The gene
Basic information
- Locus tag: BSU38750
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Ubiquinol-8 + O2 = Ubiquinone-8 + H2O (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): contains an iron-sulfur cluster
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P94365
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ankita Puri-Taneja, Matthew Schau, Yinghua Chen, F Marion Hulett
Regulators of the Bacillus subtilis cydABCD operon: identification of a negative regulator, CcpA, and a positive regulator, ResD.
J Bacteriol: 2007, 189(9);3348-58
[PubMed:17322317]
[WorldCat.org]
[DOI]
(P p)
Jonas T Larsson, Annika Rogstam, Claes von Wachenfeldt
Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 10);3323-3335
[PubMed:16207915]
[WorldCat.org]
[DOI]
(P p)
Matthew Schau, Yinghua Chen, F Marion Hulett
Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon.
J Bacteriol: 2004, 186(14);4585-95
[PubMed:15231791]
[WorldCat.org]
[DOI]
(P p)
N Azarkina, S Siletsky, V Borisov, C von Wachenfeldt, L Hederstedt, A A Konstantinov
A cytochrome bb'-type quinol oxidase in Bacillus subtilis strain 168.
J Biol Chem: 1999, 274(46);32810-7
[PubMed:10551842]
[WorldCat.org]
[DOI]
(P p)
L Winstedt, K Yoshida, Y Fujita, C von Wachenfeldt
Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon.
J Bacteriol: 1998, 180(24);6571-80
[PubMed:9852001]
[WorldCat.org]
[DOI]
(P p)