Difference between revisions of "CpgA"
| Line 56: | Line 56: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * forms curly cells {{PubMed|22544754}} | ||
=== Database entries === | === Database entries === | ||
| Line 64: | Line 65: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 84: | Line 83: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | * '''Modification:''' phosphorylated on | + | * '''Modification:''' |
| + | ** ''in vitro'' phosphorylated on Thr-166 by [[PrkC]], dephosphorylated by [[PrpC]] {{PubMed|22544754,19246764}} | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 91: | Line 91: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** binds the 30S subunit of the ribosome {{PubMed|22544754}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 146: | Line 147: | ||
<pubmed> 19575570 </pubmed> | <pubmed> 19575570 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed> 15223319, 16485133, 14747714,, 18344364, 19246764, 19246764, 15828870, 17005971, 18344364, 18984160 </pubmed> | + | <pubmed> 15223319, 16485133, 14747714,22544754, 18344364, 19246764, 19246764, 15828870, 17005971, 18344364, 18984160 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:44, 6 May 2012
- Description: GTPase, activity stimulated by ribosomes, may be involved in ribosome maturation
| Gene name | cpgA |
| Synonyms | yloQ |
| Essential | no |
| Product | GTPase |
| Function | ribosome assembly, coordination of peptidoglycan deposition in the cell wall |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 33 kDa, 4.743 |
| Gene length, protein length | 894 bp, 298 aa |
| Immediate neighbours | prkC, rpe |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 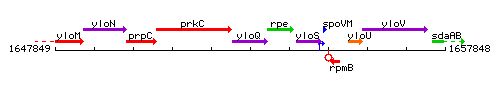
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed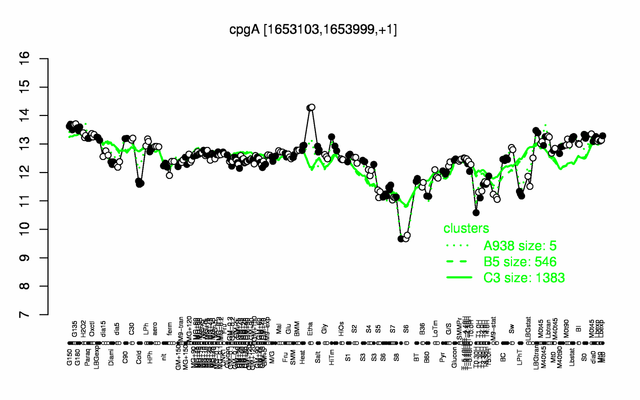
| |
Contents
Categories containing this gene/protein
cell wall/ other, translation, GTP-binding proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15780
Phenotypes of a mutant
- forms curly cells PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: engC GTPase domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- binds the 30S subunit of the ribosome PubMed
Database entries
- Structure: 1T9H
- UniProt: O34530
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Tony Wilkinson, York University, U.K. homepage
Your additional remarks
References
Reviews
Original publications
Frédérique Pompeo, Céline Freton, Catherine Wicker-Planquart, Christophe Grangeasse, Jean-Michel Jault, Anne Galinier
Phosphorylation of CpgA protein enhances both its GTPase activity and its affinity for ribosome and is crucial for Bacillus subtilis growth and morphology.
J Biol Chem: 2012, 287(25);20830-8
[PubMed:22544754]
[WorldCat.org]
[DOI]
(I p)
Cédric Absalon, Michal Obuchowski, Edwige Madec, Delphine Delattre, I Barry Holland, Simone J Séror
CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 3);932-943
[PubMed:19246764]
[WorldCat.org]
[DOI]
(P p)
Ishita M Shah, Maria-Halima Laaberki, David L Popham, Jonathan Dworkin
A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments.
Cell: 2008, 135(3);486-96
[PubMed:18984160]
[WorldCat.org]
[DOI]
(I p)
Cédric Absalon, Kassem Hamze, Didier Blanot, Claude Frehel, Rut Carballido-Lopez, Barry I Holland, Jean van Heijenoort, Simone J Séror
The GTPase CpgA is implicated in the deposition of the peptidoglycan sacculus in Bacillus subtilis.
J Bacteriol: 2008, 190(10);3786-90
[PubMed:18344364]
[WorldCat.org]
[DOI]
(I p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)
Lionel Cladière, Kassem Hamze, Edwige Madec, Vladimir M Levdikov, Anthony J Wilkinson, I Barry Holland, Simone J Séror
The GTPase, CpgA(YloQ), a putative translation factor, is implicated in morphogenesis in Bacillus subtilis.
Mol Genet Genomics: 2006, 275(4);409-20
[PubMed:16485133]
[WorldCat.org]
[DOI]
(P p)
Tracey L Campbell, Denis M Daigle, Eric D Brown
Characterization of the Bacillus subtilis GTPase YloQ and its role in ribosome function.
Biochem J: 2005, 389(Pt 3);843-52
[PubMed:15828870]
[WorldCat.org]
[DOI]
(I p)
Vladimir M Levdikov, Elena V Blagova, James A Brannigan, Lionel Cladière, Alfred A Antson, Michail N Isupov, Simone J Séror, Anthony J Wilkinson
The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability.
J Mol Biol: 2004, 340(4);767-82
[PubMed:15223319]
[WorldCat.org]
[DOI]
(P p)
Lionel Cladière, Elena Blagova, Vladimir M Levdikov, James A Brannigan, Simone J Séror, Anthony J Wilkinson
Crystallization of YloQ, a GTPase of unknown function essential for Bacillus subtilis viability.
Acta Crystallogr D Biol Crystallogr: 2004, 60(Pt 2);329-30
[PubMed:14747714]
[WorldCat.org]
[DOI]
(P p)