Difference between revisions of "YjmC"
| Line 24: | Line 24: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yjmC_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yjmC_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yjmC_1303423_1304436_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:yjmC_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/> | <br/><br/> | ||
Revision as of 07:37, 19 April 2012
- Description: unknown, may be involved in galacturonate utilization
| Gene name | yjmC |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 36 kDa, 5.669 |
| Gene length, protein length | 1011 bp, 337 aa |
| Immediate neighbours | yjmB, yjmD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed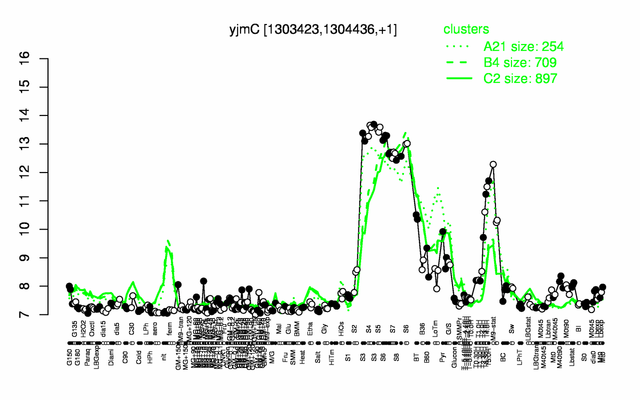
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, sporulation proteins
This gene is a member of the following regulons
CcpA regulon, ExuR regulon, SigE regulon
The gene
Basic information
- Locus tag: BSU12320
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: LDH2/MDH2 oxidoreductase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: O34736
- KEGG entry: [3]
- E.C. number:
Additional information
The gene is annotated in KEGG as an ortholog of malate dehydrogenase EC 1.1.1.37. but it is marked as “uncharacterized oxidoreductase” (EC 1.1.1.-) in Swiss-Prot. In MetaCyc it is marked as “similar to malate dehydrogenase”. As the paper by Mekjian et al. PubMed suggests this gene is more likely to be involved in the glucuronate pathway (for which EC 1.1.1.37 is not a member), no literature/experimental evidence supporting the annotation is available. PubMed
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References