Difference between revisions of "YtcA"
| Line 98: | Line 98: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ytcA_3155725_3157011_1 ytcA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 104: | Line 106: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 16:00, 16 April 2012
- Description: similar to to NDP-sugar dehydrogenase
| Gene name | ytcA |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 46 kDa, 5.943 |
| Gene length, protein length | 1284 bp, 428 aa |
| Immediate neighbours | ytdA, ytcB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 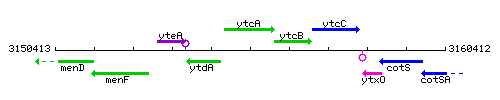
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30860
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O34862
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: