Difference between revisions of "PhoA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 100: | Line 100: | ||
* '''Operon:''' ''phoA'' {{PubMed|16291680}} | * '''Operon:''' ''phoA'' {{PubMed|16291680}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=phoA_1017083_1018468_-1 phoA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' SigA {{PubMed|8113174}}, [[SigG]] {{PubMed|15699190}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 16:50, 12 April 2012
- Description: alkaline phosphatase A
| Gene name | phoA |
| Synonyms | phoAIV |
| Essential | no |
| Product | alkaline phosphatase A |
| Function | aquisition of phosphate upon phosphoate starvation |
| MW, pI | 50 kDa, 9.926 |
| Gene length, protein length | 1383 bp, 461 aa |
| Immediate neighbours | spoVR, lytE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 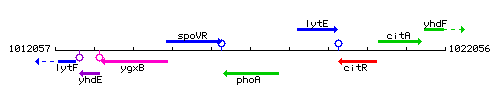
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU09410
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: A phosphate monoester + H2O = an alcohol + phosphate (according to Swiss-Prot)
- Protein family: alkaline phosphatase family (according to Swiss-Prot)
- Paralogous protein(s): PhoB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: P19406
- KEGG entry: [3]
- E.C. number: 3.1.3.1
Additional information
Expression and regulation
- Operon: phoA PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Marion Hulett, University of Illinois at Chicago, USA Homepage
Your additional remarks
References
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Soo-Keun Choi, Milton H Saier
Regulation of pho regulon gene expression by the carbon control protein A, CcpA, in Bacillus subtilis.
J Mol Microbiol Biotechnol: 2005, 10(1);40-50
[PubMed:16491025]
[WorldCat.org]
[DOI]
(P p)
Nicholas E E Allenby, Nicola O'Connor, Zoltán Prágai, Alan C Ward, Anil Wipat, Colin R Harwood
Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis.
J Bacteriol: 2005, 187(23);8063-80
[PubMed:16291680]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
H Antelmann, C Scharf, M Hecker
Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis.
J Bacteriol: 2000, 182(16);4478-90
[PubMed:10913081]
[WorldCat.org]
[DOI]
(P p)
W Liu, Y Qi, F M Hulett
Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity.
Mol Microbiol: 1998, 28(1);119-30
[PubMed:9593301]
[WorldCat.org]
[DOI]
(P p)
F M Hulett, J Lee, L Shi, G Sun, R Chesnut, E Sharkova, M F Duggan, N Kapp
Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis.
J Bacteriol: 1994, 176(5);1348-58
[PubMed:8113174]
[WorldCat.org]
[DOI]
(P p)