Difference between revisions of "YacO"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 100: | Line 100: | ||
* '''Operon:''' ''[[gltX]]-[[cysE]]-[[cysS]]-[[mrnC]]-[[yacO]]-[[yacP]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/7510287 PubMed] | * '''Operon:''' ''[[gltX]]-[[cysE]]-[[cysS]]-[[mrnC]]-[[yacO]]-[[yacP]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/7510287 PubMed] | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rlmB_115269_116018_1 yacO] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/7510287 PubMed] | ||
* '''Regulation:''' repression by cysteine ([[T-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/7510287 PubMed] | * '''Regulation:''' repression by cysteine ([[T-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/7510287 PubMed] | ||
Revision as of 11:11, 12 April 2012
- Description: putative 23S rRNA methyltransferase
| Gene name | yacO |
| Synonyms | |
| Essential | no |
| Product | putative 23S rRNA methyltransferase |
| Function | unknown |
| Metabolic function and regulation of this protein in SubtiPathways: tRNA charging | |
| MW, pI | 27 kDa, 8.633 |
| Gene length, protein length | 747 bp, 249 aa |
| Immediate neighbours | mrnC, yacP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 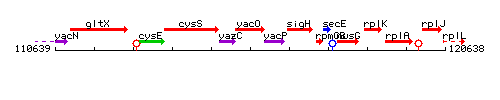
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00960
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: RNA methyltransferase trmH family (according to Swiss-Prot)
- Paralogous protein(s): YsgA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q06753
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism: T-box, overlaps a transcription terminator upstream of cysE: transcription antitermination PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ana Gutiérrez-Preciado, Tina M Henkin, Frank J Grundy, Charles Yanofsky, Enrique Merino
Biochemical features and functional implications of the RNA-based T-box regulatory mechanism.
Microbiol Mol Biol Rev: 2009, 73(1);36-61
[PubMed:19258532]
[WorldCat.org]
[DOI]
(I p)