Difference between revisions of "SipW"
(→Reviews) |
|||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || [[biofilm formation]] | |style="background:#ABCDEF;" align="center"|'''Function''' || [[biofilm formation]] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SipW SipW] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/wiki/index.php/Protein_secretion Protein secretion]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/wiki/index.php/Protein_secretion Protein secretion]''' | ||
| Line 65: | Line 67: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
** important for [[biofilm formation]] on a solid surface, but not required at an air-liquid interface {{PubMed|22328672}} | ** important for [[biofilm formation]] on a solid surface, but not required at an air-liquid interface {{PubMed|22328672}} | ||
| − | ** Cleavage of hydrophobic, N-terminal signal or leader sequences from [[TasA]] and [[TapA]] | + | ** Cleavage of hydrophobic, N-terminal signal or leader sequences from [[TasA]] and [[TapA]] {{PubMed|10049401,10559173}} |
* '''Protein family:''' peptidase S26B family (according to Swiss-Prot) | * '''Protein family:''' peptidase S26B family (according to Swiss-Prot) | ||
| Line 84: | Line 86: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[SipW]]-[[TapA]] {{PubMed|10559173}} | ||
| + | ** [[SipW]]-[[TasA]] {{PubMed|10049401}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| − | ** membrane | + | ** membrane {{PubMed|22328672}} |
=== Database entries === | === Database entries === | ||
| Line 155: | Line 159: | ||
[http://www.ncbi.nlm.nih.gov/pubmed/21856853 PubMed:21856853] | [http://www.ncbi.nlm.nih.gov/pubmed/21856853 PubMed:21856853] | ||
| − | <pubmed>18047568,15175311,16430695,10368135,9694797,18430133,18647168,10464223,17720793, 20351052 10827084 21477127 </pubmed> | + | <pubmed>18047568,15175311,16430695,10368135,9694797,18430133,18647168,10464223,17720793, 20351052 10827084 21477127 10559173 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:29, 23 March 2012
- Description: bifunctional signal peptidase I that controls surface-adhered biofilm formation and processes TasA and TapA
| Gene name | sipW |
| Synonyms | yqhE |
| Essential | no |
| Product | signal peptidase I |
| Function | biofilm formation |
| Interactions involving this protein in SubtInteract: SipW | |
| Regulation of this protein in SubtiPathways: Biofilm, Protein secretion | |
| MW, pI | 20 kDa, 5.494 |
| Gene length, protein length | 570 bp, 190 aa |
| Immediate neighbours | tasA, tapA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 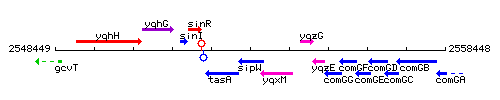
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
protein secretion, biofilm formation, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24630
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- important for biofilm formation on a solid surface, but not required at an air-liquid interface PubMed
- Cleavage of hydrophobic, N-terminal signal or leader sequences from TasA and TapA PubMed
- Protein family: peptidase S26B family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- membrane PubMed
Database entries
- Structure:
- UniProt: P54506
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Beth Lazazzera, Los Angeles, USA
Your additional remarks
References
Reviews
Additional reviews: PubMed
Original publications
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947
Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hübner S, Stülke J A Novel Factor Controlling Bistability in Bacillus subtilis: The YmdB Protein Affects Flagellin Expression and Biofilm Formation. J Bacteriol.: 2011, 193(21):5997-6007. PubMed:21856853