Difference between revisions of "PdaC"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' peptidoglycan deacetylase C <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || polysaccharide deacetylase C |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || cell wall modification |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 53 kDa, 9.105 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 53 kDa, 9.105 | ||
| Line 42: | Line 42: | ||
* '''Locus tag:''' BSU12100 | * '''Locus tag:''' BSU12100 | ||
| − | |||
| − | |||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * sensitive to lysozyme treatment {{PubMed|22277649}} | ||
=== Database entries === | === Database entries === | ||
| Line 54: | Line 53: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 63: | Line 60: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** deacetylates N-acetylmuramic acid (MurNAc) but not GlcNAc from peptidoglycan {{PubMed|22277649}} | ||
* '''Protein family:''' polysaccharide deacetylase family (according to Swiss-Prot) | * '''Protein family:''' polysaccharide deacetylase family (according to Swiss-Prot) | ||
| Line 71: | Line 69: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | + | ** K(m)=4.8 mM and k(cat)=0.32 s(-1) (toward ''B. subtilis'' peptidoglycan) {{PubMed|22277649}} | |
* '''Domains:''' | * '''Domains:''' | ||
| Line 128: | Line 126: | ||
=References= | =References= | ||
| − | <pubmed>17878218,17581128, | + | <pubmed>17878218,17581128, 22277649</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:25, 27 January 2012
- Description: peptidoglycan deacetylase C
| Gene name | yjeA |
| Synonyms | |
| Essential | no |
| Product | polysaccharide deacetylase C |
| Function | cell wall modification |
| MW, pI | 53 kDa, 9.105 |
| Gene length, protein length | 1401 bp, 467 aa |
| Immediate neighbours | cotT, yjfA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 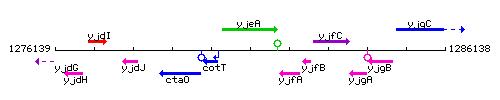
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12100
Phenotypes of a mutant
- sensitive to lysozyme treatment PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- deacetylates N-acetylmuramic acid (MurNAc) but not GlcNAc from peptidoglycan PubMed
- Protein family: polysaccharide deacetylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- K(m)=4.8 mM and k(cat)=0.32 s(-1) (toward B. subtilis peptidoglycan) PubMed
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O34798
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Kaori Kobayashi, I Putu Sudiarta, Takeko Kodama, Tatsuya Fukushima, Katsutoshi Ara, Katsuya Ozaki, Junichi Sekiguchi
Identification and characterization of a novel polysaccharide deacetylase C (PdaC) from Bacillus subtilis.
J Biol Chem: 2012, 287(13);9765-9776
[PubMed:22277649]
[WorldCat.org]
[DOI]
(I p)
Ka-Lun Ng, Chui-Chi Lam, Zhibiao Fu, Yi-Fan Han, Karl W K Tsim, Wan-Keung R Wong
Cloning and characterization of the yjeA gene, encoding a novel deoxyribonuclease, from Bacillus subtilis.
J Biochem: 2007, 142(5);647-54
[PubMed:17878218]
[WorldCat.org]
[DOI]
(P p)
Paola Bisicchia, David Noone, Efthimia Lioliou, Alistair Howell, Sarah Quigley, Thomas Jensen, Hanne Jarmer, Kevin M Devine
The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis.
Mol Microbiol: 2007, 65(1);180-200
[PubMed:17581128]
[WorldCat.org]
[DOI]
(P p)