Difference between revisions of "RsbT"
(→Original Articles) |
|||
| Line 47: | Line 47: | ||
* '''Locus tag:''' BSU04690 | * '''Locus tag:''' BSU04690 | ||
| + | |||
| + | [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rsbT_520606_521007_1 Expression] | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 14:13, 24 January 2012
- Description: PP2C activator, protein serine kinase, phosphorylates RsbS, part of the stressosome
| Gene name | rsbT |
| Synonyms | ycxT |
| Essential | no |
| Product | PP2C activator, protein serine kinase |
| Function | control of SigB activity |
| Interactions involving this protein in SubtInteract: RsbT | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 14 kDa, 6.587 |
| Gene length, protein length | 399 bp, 133 aa |
| Immediate neighbours | rsbS, rsbU |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 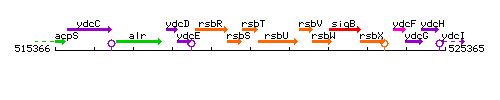
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
protein modification, sigma factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04690
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3VY9 (complete stressosome)
- UniProt: P42411
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
- Rick Lewis, Newcastle, UK homepage
Your additional remarks
References
Reviews
Original Articles
Additional publications: PubMed
Locke JC, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB Stochastic pulse regulation in bacterial stress response. Science. 2011 334:366-369. PubMed:21979936