Difference between revisions of "SsrA"
(→Expression and regulation) |
|||
| Line 60: | Line 60: | ||
* '''Operon:''' | * '''Operon:''' | ||
| + | ** ''[[secG]]-[[yvaK]]-[[rnr]]-[[smpB]]-[[ssrA]]'' {{PubMed|17369301}} | ||
| + | ** ''[[yvaK]]-[[rnr]]-[[smpB]]-[[ssrA]]'' {{PubMed|17369301}} | ||
| + | ** ''[[smpB]]-[[ssrA]]'' {{PubMed|17369301}} | ||
| + | ** ''[[ssrA]]'' {{PubMed|17369301}} | ||
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| Line 67: | Line 71: | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 07:25, 3 February 2011
- Description: tmRNA
| Gene name | ssrA |
| Synonyms | |
| Essential | no |
| Product | tmRNA |
| Function | trans-translation |
| Gene length | bp |
| Immediate neighbours | yvaG, smpB |
| Gene sequence (+200bp) | |
Genetic context 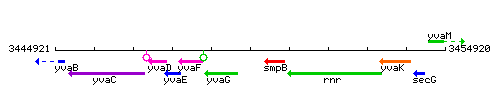
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
- Structure:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP323 (cat), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/RNA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews
Janine Kirstein, Noël Molière, David A Dougan, Kürşad Turgay
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.
Nat Rev Microbiol: 2009, 7(8);589-99
[PubMed:19609260]
[WorldCat.org]
[DOI]
(I p)
Jeffrey H Withey, David I Friedman
A salvage pathway for protein structures: tmRNA and trans-translation.
Annu Rev Microbiol: 2003, 57;101-23
[PubMed:12730326]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Hiromi Ujiie, Tomoko Matsutani, Hisashi Tomatsu, Ai Fujihara, Chisato Ushida, Yasuhiko Miwa, Yasutaro Fujita, Hyouta Himeno, Akira Muto
Trans-translation is involved in the CcpA-dependent tagging and degradation of TreP in Bacillus subtilis.
J Biochem: 2009, 145(1);59-66
[PubMed:18977770]
[WorldCat.org]
[DOI]
(I p)
Ji-Hyun Shin, Chester W Price
The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures.
J Bacteriol: 2007, 189(10);3729-37
[PubMed:17369301]
[WorldCat.org]
[DOI]
(P p)
Marc A B Kolkman, Eugenio Ferrari
The fate of extracellular proteins tagged by the SsrA system of Bacillus subtilis.
Microbiology (Reading): 2004, 150(Pt 2);427-436
[PubMed:14766921]
[WorldCat.org]
[DOI]
(P p)
T Wiegert, W Schumann
SsrA-mediated tagging in Bacillus subtilis.
J Bacteriol: 2001, 183(13);3885-9
[PubMed:11395451]
[WorldCat.org]
[DOI]
(P p)
A W Karzai, E D Roche, R T Sauer
The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue.
Nat Struct Biol: 2000, 7(6);449-55
[PubMed:10881189]
[WorldCat.org]
[DOI]
(P p)