Difference between revisions of "Sandbox"
(Removing all content from page) |
|||
| Line 1: | Line 1: | ||
| + | * '''Description:''' transcriptional repressor of gluconeogenetic genes and of [[sr1]]. repression in the presence of glucose <br/><br/> | ||
| + | {| align="right" border="1" cellpadding="2" | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| + | |''ccpN'' | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''yqzB '' | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional regulator | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"|'''Function''' || repressor of genes involved in gluconeogenesis (''[[gapB]]'','' [[pckA]]'') and of ''[[sr1]]'' | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 23.4 kDa, 7.22 | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 636 bp, 212 amino acids | ||
| + | |- | ||
| + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[glyS]]'', ''[[yqfL]]'' | ||
| + | |- | ||
| + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ccpN_nucleotide.txt Gene sequence (+200bp) ]''' | ||
| + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ccpN_protein.txt Protein sequence]''' | ||
| + | |- | ||
| + | |colspan="2" | '''Genetic context''' <br/> [[Image:yqzB_context.gif]] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | __TOC__ | ||
| + | |||
| + | <br/><br/> | ||
| + | |||
| + | |||
| + | =The gene= | ||
| + | |||
| + | === Basic information === | ||
| + | |||
| + | * '''Coordinates:''' 2604195 - 2604830 | ||
| + | |||
| + | ===Phenotypes of a mutant === | ||
| + | Impaired growth on glucose due to re-routing of carbon from glycolysis to the pentose phosphate pathway [http://www.ncbi.nlm.nih.gov/sites/entrez/18586936 PubMed] | ||
| + | |||
| + | === Database entries === | ||
| + | |||
| + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ccpN-yqfL.html] | ||
| + | |||
| + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG13768] | ||
| + | |||
| + | === Additional information=== | ||
| + | |||
| + | |||
| + | |||
| + | === Basic information/ Evolution === | ||
| + | |||
| + | * '''Catalyzed reaction/ biological activity:''' transcription repression of the ''[[gapB]]'', ''[[pckA]]'', and ''[[sr1]]'' genes in the presence of glucose [http://www.ncbi.nlm.nih.gov/sites/entrez/2548995 PubMed] | ||
| + | |||
| + | * '''Protein family:''' | ||
| + | |||
| + | * '''Paralogous protein(s):''' | ||
| + | |||
| + | === Extended information on the protein === | ||
| + | |||
| + | * '''Kinetic information:''' | ||
| + | |||
| + | * '''Domains:''' | ||
| + | |||
| + | * '''Modification:''' | ||
| + | |||
| + | * '''Cofactor(s):''' | ||
| + | |||
| + | * '''Effectors of protein activity:''' | ||
| + | ** [[YqfL]] (negative effector of CcpN activity) [http://www.ncbi.nlm.nih.gov/sites/entrez/15720552 PubMed] | ||
| + | ** ATP enhances CcpN-dependent repression, ADP counteracts the repressing activity of CcpN [http://www.ncbi.nlm.nih.gov/sites/entrez/18511073 PubMed] | ||
| + | |||
| + | * '''Interactions:''' | ||
| + | |||
| + | * '''Localization:''' | ||
| + | |||
| + | === Database entries === | ||
| + | |||
| + | * '''Structure:''' | ||
| + | |||
| + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O34994] | ||
| + | |||
| + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU25250] | ||
| + | |||
| + | === Additional information=== | ||
| + | |||
| + | |||
| + | =Expression and regulation= | ||
| + | |||
| + | * '''Operon:''' ''[[ccpN]]''-''[[yqfL]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/15720552 PubMed] | ||
| + | |||
| + | * '''Sigma factor:''' | ||
| + | |||
| + | * '''Regulation:''' constitutively expressed [http://www.ncbi.nlm.nih.gov/sites/entrez/15720552 PubMed] | ||
| + | |||
| + | * '''Additional information:''' | ||
| + | |||
| + | =Biological materials = | ||
| + | |||
| + | * '''Mutant:''' DB104 ''ccpN''::[[cat]], available in [[Sabine Brantl]] lab | ||
| + | |||
| + | * '''Expression vector:''' | ||
| + | |||
| + | * '''lacZ fusion:''' | ||
| + | |||
| + | * '''GFP fusion:''' | ||
| + | |||
| + | * '''Antibody:''' available in [[Sabine Brantl]] lab | ||
| + | |||
| + | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Stephane Aymerich |Stephane Aymerich]], Microbiology and Molecular Genetics, INRA Paris-Grignon, France | ||
| + | |||
| + | [[Sabine Brantl |Sabine Brantl]], Bacterial Genetics, Friedrich-Schiller-University of Jena, Germany [http://www.uni-jena.de/AG_Bakteriengenetik_(_Doz__Dr__Sabine_Brantl_).html homepage] | ||
| + | |||
| + | =Your additional remarks= | ||
| + | |||
| + | =References= | ||
| + | |||
| + | |||
| + | # Servant et al. (2005) CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of ''Bacillus subtilis'' gluconeogenic genes. Mol. Microbiol. 55: 1435-1451. [http://www.ncbi.nlm.nih.gov/sites/entrez/15720552 PubMed] | ||
| + | # Licht et al. (2005) Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in ''Bacillus subtilis''. Mol. Microbiol. 58: 189-206. [http://www.ncbi.nlm.nih.gov/sites/entrez/16164558 PubMed] | ||
| + | # Licht & Brantl (2006) Transcriptional repressor CcpN from ''Bacillus subtilis'' compensates asymmetric contact distribution by cooperative binding. J. Mol. Biol. 364: 434-448. [http://www.ncbi.nlm.nih.gov/sites/entrez/17011578 PubMed] | ||
| + | # Licht et al. (2008) Identification of ligands affecting the activity of the transcriptional repressor CcpN from ''Bacillus subtilis''. J. Mol. Biol. 380: 17-30. [http://www.ncbi.nlm.nih.gov/sites/entrez/18511073 PubMed] | ||
| + | # Tännler et al. (2008) CcpN controls central carbon fluxes in ''Bacillus subtilis''. J. Bacteriol. 190: 6178-6187. [http://www.ncbi.nlm.nih.gov/sites/entrez/18586936 PubMed] | ||
Revision as of 15:23, 10 February 2009
- Description: transcriptional repressor of gluconeogenetic genes and of sr1. repression in the presence of glucose
| Gene name | ccpN |
| Synonyms | yqzB |
| Essential | no |
| Product | transcriptional regulator |
| Function | repressor of genes involved in gluconeogenesis (gapB, pckA) and of sr1 |
| MW, pI | 23.4 kDa, 7.22 |
| Gene length, protein length | 636 bp, 212 amino acids |
| Immediate neighbours | glyS, yqfL |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 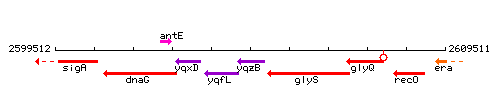
| |
Contents
The gene
Basic information
- Coordinates: 2604195 - 2604830
Phenotypes of a mutant
Impaired growth on glucose due to re-routing of carbon from glycolysis to the pentose phosphate pathway PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription repression of the gapB, pckA, and sr1 genes in the presence of glucose PubMed
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
Additional information
Expression and regulation
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Additional information:
Biological materials
- Mutant: DB104 ccpN::cat, available in Sabine Brantl lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody: available in Sabine Brantl lab
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Sabine Brantl, Bacterial Genetics, Friedrich-Schiller-University of Jena, Germany homepage
Your additional remarks
References
- Servant et al. (2005) CcpN (YqzB), a novel regulator for CcpA-independent catabolite repression of Bacillus subtilis gluconeogenic genes. Mol. Microbiol. 55: 1435-1451. PubMed
- Licht et al. (2005) Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol. Microbiol. 58: 189-206. PubMed
- Licht & Brantl (2006) Transcriptional repressor CcpN from Bacillus subtilis compensates asymmetric contact distribution by cooperative binding. J. Mol. Biol. 364: 434-448. PubMed
- Licht et al. (2008) Identification of ligands affecting the activity of the transcriptional repressor CcpN from Bacillus subtilis. J. Mol. Biol. 380: 17-30. PubMed
- Tännler et al. (2008) CcpN controls central carbon fluxes in Bacillus subtilis. J. Bacteriol. 190: 6178-6187. PubMed