Difference between revisions of "SndA"
(→References) |
(→References) |
||
| Line 125: | Line 125: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|15272571}} | |
| − | <pubmed>16109943,15668000, | + | <pubmed>16109943,15668000,</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 21:48, 6 December 2010
- Description: unknown
| Gene name | ytnL |
| Synonyms | hipO |
| Essential | no |
| Product | unknown |
| Function | unknown |
| Metabolic function and regulation of this protein in SubtiPathways: Riboflavin / FAD | |
| MW, pI | 45 kDa, 5.545 |
| Gene length, protein length | 1248 bp, 416 aa |
| Immediate neighbours | ytnM, ribR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 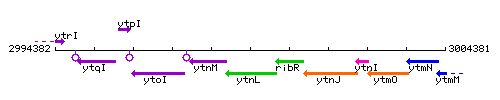
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU29290
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Categories containing this gene/protein
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase M20 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O34980
- KEGG entry: [3]
- E.C. number:
Additional information
The gene was annotated in KEGG and MetaCyc as hippurate hydrolase EC 3.5.1.32. In Swiss-Prot it is marked as “uncharacterized hydrolase”. No literature/experimental evidence supporting ththeannotation was removed as of November 2009. PubMed
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Pierre Burguière, Juliette Fert, Isabelle Guillouard, Sandrine Auger, Antoine Danchin, Isabelle Martin-Verstraete
Regulation of the Bacillus subtilis ytmI operon, involved in sulfur metabolism.
J Bacteriol: 2005, 187(17);6019-30
[PubMed:16109943]
[WorldCat.org]
[DOI]
(P p)
Irina M Solovieva, Rimma A Kreneva, Lubov Errais Lopes, Daniel A Perumov
The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product.
FEMS Microbiol Lett: 2005, 243(1);51-8
[PubMed:15668000]
[WorldCat.org]
[DOI]
(P p)