Difference between revisions of "SigA"
(→References) |
|||
| Line 128: | Line 128: | ||
=References= | =References= | ||
| − | <pubmed> 19680289 20817769 , | + | <pubmed> 19680289 20817769 ,,</pubmed> |
| − | <pubmed>, | + | <pubmed>2997585,9658000,3127379,</pubmed> |
| − | =2= | + | ==2== |
| − | <pubmed> 12486038,10556308,8107087, 14762002,7918363,8071196,7630711,9185571,9287022, 14679239, 14976210 19735077, </pubmed> | + | <pubmed> 12486038,10556308,8107087, 14762002,7918363,8071196,7630711,9185571,9287022, 14679239, 14976210 19735077, 10438769,11535782,7599136, 9144176</pubmed> |
Additional publication: {{PubMed|20935043}} | Additional publication: {{PubMed|20935043}} | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:26, 17 October 2010
- Description: RNA polymerase major sigma factor SigA
| Gene name | sigA |
| Synonyms | rpoD, crsA |
| Essential | yes PubMed |
| Product | RNA polymerase major sigma factor SigA |
| Function | transcription |
| MW, pI | 42 kDa, 4.634 |
| Gene length, protein length | 1113 bp, 371 aa |
| Immediate neighbours | cccA, antE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 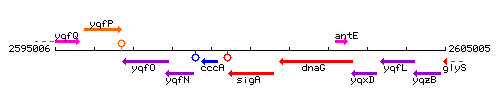
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25200
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P06224
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
2
Additional publication: PubMed