Difference between revisions of "SlrR"
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcriptional activator of competence development and sporulation genes, represses [[SigD]]-dependent flagellar genes, antagonist of [[SlrA]] and [[SinR]]<br/><br/> | + | * '''Description:''' transcriptional activator of competence development and sporulation genes, represses [[SigD]]-dependent flagellar genes, antagonist of [[SlrA]] and [[SinR]], has LexA-like autocleavage activity<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 54: | Line 54: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' binding and concomintant inactivation of [[SlrA]] results in release of [[SinR]] from the [[SinR]]-[[SlrA]] complex and subsequent induction of [[SinR]]-controlled genes and operons for biofilm formation {{PubMed|19788541}} | + | * '''Catalyzed reaction/ biological activity:''' |
| + | ** binding and concomintant inactivation of [[SlrA]] results in release of [[SinR]] from the [[SinR]]-[[SlrA]] complex and subsequent induction of [[SinR]]-controlled genes and operons for biofilm formation {{PubMed|19788541}} | ||
| + | ** repression of transcription of ''[[lytA]]-[[lytB]]-[[lytC]]'' and ''[[lytF]]'' {{PubMed|20351052}} | ||
| + | ** autocleavage {{PubMed|20923420}} | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| Line 67: | Line 70: | ||
* '''Modification:''' | * '''Modification:''' | ||
| + | ** subject to self-cleavage via a LexA-like autopeptidase, this involves [[ClpC]] {{PubMed|20923420}} | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 72: | Line 76: | ||
* '''Effectors of protein activity:''' interaction with [[SinR]] triggers binding of SlrR to the promoters of ''[[lytA]]-[[lytB]]-[[lytC]]'' and ''[[lytF]]'', resulting in their repression {{PubMed|20351052}} | * '''Effectors of protein activity:''' interaction with [[SinR]] triggers binding of SlrR to the promoters of ''[[lytA]]-[[lytB]]-[[lytC]]'' and ''[[lytF]]'', resulting in their repression {{PubMed|20351052}} | ||
| − | * '''Interactions:''' [[SlrA]]-[[SlrR]] {{PubMed|19788541}} | + | * '''Interactions:''' |
| + | ** [[SlrA]]-[[SlrR]] {{PubMed|19788541}} | ||
| + | ** [[SinR]]-[[SlrR]] {{PubMed|20351052}} | ||
* '''Localization:''' | * '''Localization:''' | ||
| Line 126: | Line 132: | ||
<pubmed> 20541494 </pubmed> | <pubmed> 20541494 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>18430133,18647168, 19767430, 19788541 20351052 20817675</pubmed> | + | <pubmed>18430133,18647168, 19767430, 19788541 20351052 20817675 20923420 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:17, 7 October 2010
- Description: transcriptional activator of competence development and sporulation genes, represses SigD-dependent flagellar genes, antagonist of SlrA and SinR, has LexA-like autocleavage activity
| Gene name | slrR |
| Synonyms | yveJ, slr |
| Essential | no |
| Product | transcription regulator, SlrA antagonist |
| Function | regulation of initiation of biofilm formation and of autolysis |
| Regulation of this protein in SubtiPathways: Biofilm | |
| MW, pI | 17 kDa, 9.63 |
| Gene length, protein length | 456 bp, 152 aa |
| Immediate neighbours | epsA, pnbA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 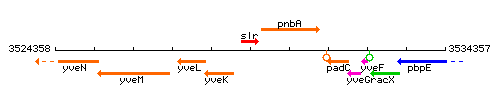
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU34380
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): SinR
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s):
- Effectors of protein activity: interaction with SinR triggers binding of SlrR to the promoters of lytA-lytB-lytC and lytF, resulting in their repression PubMed
- Localization:
Database entries
- Structure:
- UniProt: P71049
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications