Difference between revisions of "MetN"
| Line 98: | Line 98: | ||
** induced by methionine starvation ([[S-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10094622 PubMed] | ** induced by methionine starvation ([[S-box]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10094622 PubMed] | ||
| − | * '''Regulatory mechanism:''' [[S-box]]: transcription termination/ antitermination, the [[S-box]] [[riboswitch]] binds S-adenosylmethionine resulting in termination [http://www.ncbi.nlm.nih.gov/sites/entrez/10094622 PubMed] | + | * '''Regulatory mechanism:''' |
| + | ** [[S-box]]: transcription termination/ antitermination, the [[S-box]] [[riboswitch]] binds S-adenosylmethionine resulting in termination [http://www.ncbi.nlm.nih.gov/sites/entrez/10094622 PubMed] | ||
** [[CodY]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ** [[CodY]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ||
| Line 123: | Line 124: | ||
=References= | =References= | ||
| − | <pubmed>10092453,14990259,,12618455,18763711, </pubmed> | + | <pubmed>10092453,14990259,,12618455,18763711, 10094622 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:02, 28 December 2009
- Description: methionine ABC transporter (ATP-binding protein)
| Gene name | metN |
| Synonyms | yusC |
| Essential | no |
| Product | methionine ABC transporter (ATP-binding protein) |
| Function | methionine uptake |
| Metabolic function and regulation of this protein in SubtiPathways: Cys, Met & Sulfate assimilation | |
| MW, pI | 37 kDa, 7.303 |
| Gene length, protein length | 1023 bp, 341 aa |
| Immediate neighbours | metP, yusD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 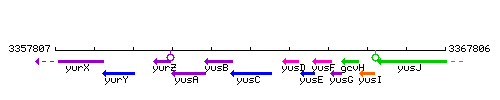
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU32750
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot), membrane PubMed
Database entries
- Structure:
- UniProt: O32169
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Marie-Françoise Hullo, Sandrine Auger, Elie Dassa, Antoine Danchin, Isabelle Martin-Verstraete
The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, D- and L-methionine.
Res Microbiol: 2004, 155(2);80-6
[PubMed:14990259]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Yoshiko Nakaura, Robert P Shivers, Hirotake Yamaguchi, Richard Losick, Yasutaro Fujita, Abraham L Sonenshein
Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis.
J Bacteriol: 2003, 185(6);1911-22
[PubMed:12618455]
[WorldCat.org]
[DOI]
(P p)
F J Grundy, T M Henkin
The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria.
Mol Microbiol: 1998, 30(4);737-49
[PubMed:10094622]
[WorldCat.org]
[DOI]
(P p)
Y Quentin, G Fichant, F Denizot
Inventory, assembly and analysis of Bacillus subtilis ABC transport systems.
J Mol Biol: 1999, 287(3);467-84
[PubMed:10092453]
[WorldCat.org]
[DOI]
(P p)