Difference between revisions of "Sda"
| Line 1: | Line 1: | ||
| − | * '''Description:''' Sda is a checkpoint protein that is upregulated in response to replication stress | + | * '''Description:''' [[Sda]] is a checkpoint protein that is upregulated in response to replication stress {{PubMed|11207367}}. [[Sda]] inhibits the autokinase activity of [[KinA]] (and likely [[KinB]] as well). This in its turn results in reduced levels of [[Spo0A|Spo0A~P]]. Thus, replication stressed cells will not initiate sporulation<br/><br/> [[Sda]] is rapidly turned over by the [[ClpP|ClpP/X]] system because of some uncharged residues at its C-terminus {{PubMed|16796683}}. |
| Line 13: | Line 13: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || developmental checkpoint protein | |style="background:#ABCDEF;" align="center"| '''Product''' || developmental checkpoint protein | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || mediates a developmental checkpoint coupling initiation of | + | |style="background:#ABCDEF;" align="center"|'''Function''' || mediates a developmental checkpoint coupling<br/>initiation of sporulation (phosphorylation of [[Spo0A]])<br/>to the function of replication initiation proteins |
| − | |||
| − | sporulation (phosphorylation of Spo0A) to the function | ||
| − | |||
| − | of replication initiation proteins | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 6 kDa, 5.605 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 6 kDa, 5.605 | ||
| Line 35: | Line 31: | ||
| − | <br/><br/> | + | <br/><br/><br/><br/> |
| Line 85: | Line 81: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3D36 3D36] (from Geobacillus stearothermophilus, complex with KinB), [http://www.rcsb.org/pdb/explore.do?structureId=1PV0 1PV0] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=3D36 3D36] (from Geobacillus stearothermophilus, complex with [[KinB]]), [http://www.rcsb.org/pdb/explore.do?structureId=1PV0 1PV0] |
* '''UniProt:''' [http://www.uniprot.org/uniprot/Q7WY62 Q7WY62] | * '''UniProt:''' [http://www.uniprot.org/uniprot/Q7WY62 Q7WY62] | ||
| Line 97: | Line 93: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''sda'' | + | * '''Operon:''' ''[[sda]]'' |
* '''Sigma factor:''' [[SigA]] | * '''Sigma factor:''' [[SigA]] | ||
Revision as of 14:36, 29 July 2009
- Description: Sda is a checkpoint protein that is upregulated in response to replication stress PubMed. Sda inhibits the autokinase activity of KinA (and likely KinB as well). This in its turn results in reduced levels of Spo0A~P. Thus, replication stressed cells will not initiate sporulation
Sda is rapidly turned over by the ClpP/X system because of some uncharged residues at its C-terminus PubMed.
| Gene name | sda |
| Synonyms | |
| Essential | no |
| Product | developmental checkpoint protein |
| Function | mediates a developmental checkpoint coupling initiation of sporulation (phosphorylation of Spo0A) to the function of replication initiation proteins |
| MW, pI | 6 kDa, 5.605 |
| Gene length, protein length | 156 bp, 52 aa |
| Immediate neighbours | yloV, sdaAA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 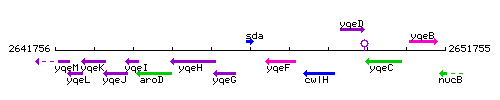
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25690
Phenotypes of a mutant
- sporulates at a higher frequency
- will sporulate in the presence of replication stress
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- UniProt: Q7WY62
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: sda
- Sigma factor: SigA
- Regulatory mechanism: Probably activated by DnaA under replication stress
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Bill Burkholder http://burkholder.stanford.edu/
Your additional remarks
References