Difference between revisions of "XlyA"
| Line 80: | Line 80: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P39800 P39800] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P39800 P39800] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU12810] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/3.5.1.28 3.5.1.28] | * '''E.C. number:''' [http://www.expasy.org/enzyme/3.5.1.28 3.5.1.28] | ||
Revision as of 05:17, 25 June 2009
- Description: N-acetylmuramoyl-L-alanine amidase
| Gene name | xlyA |
| Synonyms | |
| Essential | no |
| Product | N-acetylmuramoyl-L-alanine amidase |
| Function | PBSX prophage-mediated lysis |
| MW, pI | 31 kDa, 5.342 |
| Gene length, protein length | 891 bp, 297 aa |
| Immediate neighbours | xhlB, spoIISB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 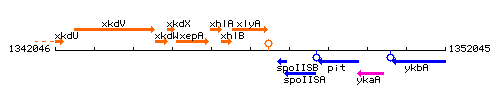
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU12810
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides (according to Swiss-Prot)
- Protein family: LysM repeat (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: secreted (according to Swiss-Prot), extracellular (no signal peptide) PubMed
Database entries
- Structure:
- Swiss prot entry: P39800
- KEGG entry: [2]
- E.C. number: 3.5.1.28
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
S Krogh, S T Jørgensen, K M Devine
Lysis genes of the Bacillus subtilis defective prophage PBSX.
J Bacteriol: 1998, 180(8);2110-7
[PubMed:9555893]
[WorldCat.org]
[DOI]
(P p)
P F Longchamp, C Mauël, D Karamata
Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-L-alanine amidase gene of prophage PBSX.
Microbiology (Reading): 1994, 140 ( Pt 8);1855-67
[PubMed:7921239]
[WorldCat.org]
[DOI]
(P p)