Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' regulation of a large regulon in response to branched-chain amino acid limitation <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''codY'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional pleiotropic repressor |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of a large regulon in response to |
| − | + | branched-chain amino acid limitation | |
| − | + | ||
| − | + | to the presence of branched-chain amino acids | |
| − | |||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|''' | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 28 kDa, 4.75 |
|- | |- | ||
| − | | | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 777 bp, 259 aa |
|- | |- | ||
| + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[clpY]]'', ''[[flgB]]'' | ||
|- | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13490]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | ||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:codY_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 31: | Line 32: | ||
__TOC__ | __TOC__ | ||
| − | <br/><br/> | + | <br/><br/><br/><br/> |
=The gene= | =The gene= | ||
| Line 37: | Line 38: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU16170 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | |||
| + | No swarming motility on B medium. [http://www.ncbi.nlm.nih.gov/sites/entrez/19202088 PubMed] | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/codV-clpQY-codY.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10968] |
=== Additional information=== | === Additional information=== | ||
| Line 56: | Line 59: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' codY family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| + | |||
| + | === Genes/ operons controlled by CodY === | ||
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 66: | Line 71: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | * '''Modification:''' phosphorylation on Ser- | + | * '''Modification:''' phosphorylation on Ser-215 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] |
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 74: | Line 79: | ||
* '''Interactions:''' | * '''Interactions:''' | ||
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasm (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2B0L 2B0L] (C-terminal DNA-binding domain), [http://www.rcsb.org/pdb/explore.do?structureId=2GX5 2GX5] (N-terminal Gaf domain) |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P39779 P39779] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU16170] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 115: | Line 120: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Linc Sonenshein|Linc Sonenshein]], Tufts University, Boston, MA, USA [http://www.tufts.edu/sackler/microbiology/faculty/sonenshein/index.html Homepage] | ||
| + | |||
| + | [[Tony Wilkinson]], York University, U.K. [http://www.york.ac.uk/depts/chem/staff/ajw.html homepage] | ||
| + | |||
| + | [[Oscar Kuipers]], University of Groningen, The Netherlands | ||
| + | [http://molgen.biol.rug.nl/molgen/index.php Homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 120: | Line 132: | ||
=References= | =References= | ||
| − | <pubmed>17218307, </pubmed> | + | <pubmed>17218307 12591885 19202088 12618455 15802253 11331605 15228537 8793880 15937175 15916605 15916606, </pubmed> |
# Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium ''Bacillus subtilis''. Mol. Cell. Proteomics 6: 697-707 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed] | # Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium ''Bacillus subtilis''. Mol. Cell. Proteomics 6: 697-707 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed] | ||
| + | # Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. (2003) Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed] | ||
| + | # Hamze et al. (2009) Identification of genes required for different stages of dendritic swarming in ''Bacillus subtilis'', with a novel role for ''phrC''. ''Microbiology'' '''155:''' 398-412. [http://www.ncbi.nlm.nih.gov/sites/entrez/19202088 PubMed] | ||
| + | # Molle V, Nakura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL: Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 2003, 185:1911-1922. [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ||
| + | # Sonenshein AL: CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 2005, 8:203-207. [http://www.ncbi.nlm.nih.gov/sites/entrez/15802253 PubMed] | ||
| + | # Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL: Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 2001, 15:1093-1103.[http://www.ncbi.nlm.nih.gov/sites/entrez/11331605 PubMed] | ||
| + | # Shivers RP, Sonenshein AL: Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 2004, 53:599-611. [http://www.ncbi.nlm.nih.gov/sites/entrez/15228537 PubMed] | ||
| + | # Serror P, Sonenshein AL: Interaction of CodY, the novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol 1996, 20:843-852. [http://www.ncbi.nlm.nih.gov/sites/entrez/8793880 PubMed] | ||
| + | # Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL: A region of Bacillus subtilis CodY protein required for interaction with DNA. J Bacteriol 2005, 187:4127-4139. [http://www.ncbi.nlm.nih.gov/sites/entrez/15937175 PubMed] | ||
| + | # Shivers RP, Sonenshein AL: Bacillus subtilis ilvB operon: an intersection of global regulons. Mol Microbiol 2005, 56:1549-1559. [http://www.ncbi.nlm.nih.gov/sites/entrez/15916605 PubMed] | ||
| + | # Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y: Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 2005, 56:1560-1573. [http://www.ncbi.nlm.nih.gov/sites/entrez/15916606 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 13:11, 8 June 2009
- Description: regulation of a large regulon in response to branched-chain amino acid limitation
| Gene name | codY |
| Synonyms | |
| Essential | no |
| Product | transcriptional pleiotropic repressor |
| Function | regulation of a large regulon in response to
branched-chain amino acid limitation to the presence of branched-chain amino acids |
| MW, pI | 28 kDa, 4.75 |
| Gene length, protein length | 777 bp, 259 aa |
| Immediate neighbours | clpY, flgB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 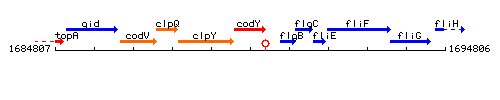
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU16170
Phenotypes of a mutant
No swarming motility on B medium. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: codY family (according to Swiss-Prot)
- Paralogous protein(s):
Genes/ operons controlled by CodY
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-215 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Swiss prot entry: P39779
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Tony Wilkinson, York University, U.K. homepage
Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed
- Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. (2003) Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. PubMed
- Hamze et al. (2009) Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC. Microbiology 155: 398-412. PubMed
- Molle V, Nakura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL: Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 2003, 185:1911-1922. PubMed
- Sonenshein AL: CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 2005, 8:203-207. PubMed
- Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL: Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 2001, 15:1093-1103.PubMed
- Shivers RP, Sonenshein AL: Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 2004, 53:599-611. PubMed
- Serror P, Sonenshein AL: Interaction of CodY, the novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol 1996, 20:843-852. PubMed
- Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL: A region of Bacillus subtilis CodY protein required for interaction with DNA. J Bacteriol 2005, 187:4127-4139. PubMed
- Shivers RP, Sonenshein AL: Bacillus subtilis ilvB operon: an intersection of global regulons. Mol Microbiol 2005, 56:1549-1559. PubMed
- Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y: Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 2005, 56:1560-1573. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed