Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' pyruvate kinase, glycolytic enzyme <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''pyk'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''pykA'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || pyruvate kinase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | |style="background:#ABCDEF;" align="center"|'''Function''' || catabolic enzyme in glycolysis | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 61,9 kDa, 4.88 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1755 bp, 585 amino acids |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[pfkA]]'', ''[[ytzA]]'' |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"|'''Hier soll was neues rein''' | |colspan="2" style="background:#FAF8CC;" align="center"|'''Hier soll was neues rein''' | ||
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:pyk_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Coordinates:''' | + | * '''Coordinates:''' 2983830 - 2985584 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | + | Unable to grow with non-PTS carbohydrates (such as glucitol or glycerol) as single carbon source. | |
=== Database entries === | === Database entries === | ||
| Line 45: | Line 45: | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/pfkA-pyk-ytzA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/pfkA-pyk-ytzA.html] | ||
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12661] |
=== Additional information=== | === Additional information=== | ||
| Line 53: | Line 53: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:'''ADP + phosphoenolpyruvate --> ATP + pyruvate, the reaction is irreversible under physiological conditions |
| − | * '''Protein family:''' | + | * '''Protein family:''' pyruvate kinase family, (C-terminal section: PEP-utilizing enzyme family) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 64: | Line 64: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | |||
| − | * '''Modification:''' | + | * '''Modification:''' phosphorylation on Ser36 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], phosphorylation on Ser536 or Ser546 [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] |
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' magnesium ion, potassium |
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' activated by PEP [http://www.ncbi.nlm.nih.gov/sites/entrez/4623707 PubMed] |
| − | * '''Interactions:''' | + | * '''Interactions:''' |
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasm [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] |
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' ''Geobacillus stearothermophilus'' [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid= | + | * '''Structure:''' 2E28 (from ''Geobacillus stearothermophilus'') [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=62183 NCBI] |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P80885] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU29180] |
| − | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.1. | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.1.40 2.7.1.40] |
=== Additional information=== | === Additional information=== | ||
| Line 91: | Line 90: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''pfkA [[pyk]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/11489127 PubMed] | + | * '''Operon:''' ''[[pfkA]] [[pyk]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/11489127 PubMed] |
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
| Line 103: | Line 102: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' GP590 (cat), available in [[Stülke]] lab |
| − | * '''Expression vector:''' | + | * '''Expression vector:''' |
| − | + | Expression in ''E. coli'', N-terminal His-tag: pGP1100 (in [[pWH844]]), available in [[Stülke]] lab | |
| − | + | ||
| + | Expression in ''B. subtilis'', native protein: pGP1411 (in [[pBQ200]]), available in [[Stülke]] lab | ||
| + | |||
| + | Expression in ''B. subtilis'', N-terminal Strep-tag: pGP1409 (in [[pGP380]]), available in [[Stülke]] lab | ||
| + | |||
| + | Expression in ''B. subtilis'', C-terminal Strep-tag: pGP1410 (in [[pGP382]]), available in [[Stülke]] lab | ||
| + | |||
| + | * '''lacZ fusion:''' see ''[[pfkA]]'' | ||
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| Line 123: | Line 129: | ||
=References= | =References= | ||
| − | # | + | # Eymann et al. (2007) Dynamics of protein phosphorylation on Ser/Thr/Tyr in ''Bacillus subtilis''. ''Proteomics'' '''7:''' 3509-3526. [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] |
| − | # | + | # Lévine et al. (2006) Analysis of the dynamic ''Bacillus subtilis'' Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. ''Proteomics'' '''6:''' 2157-2173 [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed] |
| − | # Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in ''Bacillus subtilis'': evidence for the presence of multiple levels of control of the ''gapA'' operon. Mol Microbiol 41, 409-422. [http://www.ncbi.nlm.nih.gov/sites/entrez/11489127 PubMed] | + | # Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in ''Bacillus subtilis'': evidence for the presence of multiple levels of control of the ''gapA'' operon. Mol Microbiol 41, 409-422.[http://www.ncbi.nlm.nih.gov/sites/entrez/11489127 PubMed] |
| + | # Jannière, L., Canceill, D., Suski, C., Kanga, S., Dalmais, B., Lestini, R., Monnier, A. F., Chapuis, J., Bolotin, A., Titok, M., Le Chatelier, E., and Ehrlich, S. D. (2007) Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE 2, e447. [http://www.ncbi.nlm.nih.gov/sites/entrez/17505547 PubMed] | ||
| + | # Fry, B., Zhu, T., Domach, M. M., Koepsel, R. R., Phalakornkule, C., and Ataai, M. M. (2000) Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant. Appl Env Microbiol 66: 4045-4049. [http://www.ncbi.nlm.nih.gov/sites/entrez/10966427 PubMed] | ||
| + | # Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium ''Bacillus subtilis''. Mol. Cell. Proteomics 6: 697-707 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed] | ||
Revision as of 17:18, 15 April 2009
- Description: pyruvate kinase, glycolytic enzyme
| Gene name | pyk |
| Synonyms | pykA |
| Essential | no |
| Product | pyruvate kinase |
| Function | catabolic enzyme in glycolysis |
| MW, pI | 61,9 kDa, 4.88 |
| Gene length, protein length | 1755 bp, 585 amino acids |
| Immediate neighbours | pfkA, ytzA |
| Hier soll was neues rein | |
Genetic context 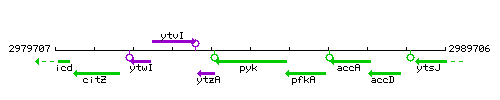
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 2983830 - 2985584
Phenotypes of a mutant
Unable to grow with non-PTS carbohydrates (such as glucitol or glycerol) as single carbon source.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:ADP + phosphoenolpyruvate --> ATP + pyruvate, the reaction is irreversible under physiological conditions
- Protein family: pyruvate kinase family, (C-terminal section: PEP-utilizing enzyme family)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s): magnesium ion, potassium
- Effectors of protein activity: activated by PEP PubMed
- Interactions:
- Localization: cytoplasm PubMed
Database entries
- Structure: 2E28 (from Geobacillus stearothermophilus) NCBI
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: 2.7.1.40
Additional information
Expression and regulation
- Sigma factor:
- Regulation: twofold induced by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP590 (cat), available in Stülke lab
- Expression vector:
Expression in E. coli, N-terminal His-tag: pGP1100 (in pWH844), available in Stülke lab
Expression in B. subtilis, native protein: pGP1411 (in pBQ200), available in Stülke lab
Expression in B. subtilis, N-terminal Strep-tag: pGP1409 (in pGP380), available in Stülke lab
Expression in B. subtilis, C-terminal Strep-tag: pGP1410 (in pGP382), available in Stülke lab
- lacZ fusion: see pfkA
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Eymann et al. (2007) Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 7: 3509-3526. PubMed
- Lévine et al. (2006) Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6: 2157-2173 PubMed
- Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol 41, 409-422.PubMed
- Jannière, L., Canceill, D., Suski, C., Kanga, S., Dalmais, B., Lestini, R., Monnier, A. F., Chapuis, J., Bolotin, A., Titok, M., Le Chatelier, E., and Ehrlich, S. D. (2007) Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE 2, e447. PubMed
- Fry, B., Zhu, T., Domach, M. M., Koepsel, R. R., Phalakornkule, C., and Ataai, M. M. (2000) Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant. Appl Env Microbiol 66: 4045-4049. PubMed
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed