Difference between revisions of "Scr"
(→References) |
|||
| Line 78: | Line 78: | ||
* '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU_MISC_RNA_2&redirect=T BSU_misc_RNA_2] | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU_MISC_RNA_2&redirect=T BSU_misc_RNA_2] | ||
| − | * '''Structure''': | + | * '''Structure''': [http://www.rcsb.org/pdb/explore.do?structureId=4WFL 4WFL] {{PubMed|25270875}} |
=== Additional information=== | === Additional information=== | ||
| Line 112: | Line 112: | ||
<pubmed> 25212246 </pubmed> | <pubmed> 25212246 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>9346318,2468993,17576666, 1372600,7506707 1383945 7538759 10224127 9677377 8662730 10222262</pubmed> | + | <pubmed>9346318,2468993,17576666, 1372600,7506707 1383945 7538759 10224127 9677377 8662730 10222262 25270875 </pubmed> |
[[Category:small RNA]] | [[Category:small RNA]] | ||
Revision as of 15:22, 4 October 2014
- Description: 4.5 S RNA
| Gene name | scr |
| Synonyms | |
| Essential | yes |
| Product | small cytoplasmic RNA (scRNA, 4.5S RNA),
signal recognition particle-like (SRP) component |
| Function | presecretory protein translocation |
| Interactions involving this RNA in SubtInteract: Scr | |
| Immediate neighbours | yaaJ, dnaX |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 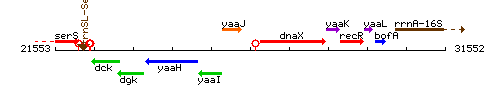
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
protein secretion, essential genes, ncRNA
This gene is a member of the following regulons
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
- the gene is essential PubMed
Database entries
- BsubCyc: BSU_misc_RNA_2
- DBTBS entry: no entry
- SubtiList entry: [1]
- Structure:
Additional information
The RNA
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
Extended information on the RNA
- Kinetic information
Database entries
- BsubCyc: BSU_misc_RNA_2
Additional information
Expression and regulation
- Sigma factor: SigA (both promoters)
- Regulation:
- Regulatory mechanism:
- Additional information: transcribed as 354 nt precursor, cleaved at the 5' end by RNase III and at the 3' site by RNase J1 to yield a 275 nt molecule, final trimming to 271 nt sc RNA by RNase PH PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References
Reviews
Original publications