Difference between revisions of "MreB"
(→Categories containing this gene/protein) |
|||
| Line 99: | Line 99: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** part of the [[cell wall biosynthetic complex]] {{PubMed|21636744,21636745}} | ** part of the [[cell wall biosynthetic complex]] {{PubMed|21636744,21636745}} | ||
| + | ** forms antiparallel double filaments {{PubMed|24843005}} | ||
** [[MreB]]-[[TufA]] {{PubMed|20133608}} | ** [[MreB]]-[[TufA]] {{PubMed|20133608}} | ||
** [[MreB]]-[[Mbl]] {{PubMed|21636744}} | ** [[MreB]]-[[Mbl]] {{PubMed|21636744}} | ||
| Line 111: | Line 112: | ||
** during logarithmic growth, [[MreB]] forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis {{PubMed|21636744}} | ** during logarithmic growth, [[MreB]] forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis {{PubMed|21636744}} | ||
** forms transverse bands as cells enter the stationary phase {{PubMed|21636744}} | ** forms transverse bands as cells enter the stationary phase {{PubMed|21636744}} | ||
| + | ** forms antiparallel double filaments {{PubMed|24843005}} | ||
** close to the inner surface of the cytoplasmic membrane [http://www.ncbi.nlm.nih.gov/sites/entrez/16950129 PubMed] | ** close to the inner surface of the cytoplasmic membrane [http://www.ncbi.nlm.nih.gov/sites/entrez/16950129 PubMed] | ||
** reports on helical structures formed by MreB {{PubMed|16950129,20566861}} seem to be misinterpretation of data {{PubMed|21636744}} | ** reports on helical structures formed by MreB {{PubMed|16950129,20566861}} seem to be misinterpretation of data {{PubMed|21636744}} | ||
| Line 176: | Line 178: | ||
==Localization== | ==Localization== | ||
| − | <pubmed> 17064365, 20566861 21636744 21636745 22069484 23783036 24010660 </pubmed> | + | <pubmed> 17064365, 20566861 21636744 21636745 22069484 23783036 24010660 24843005 </pubmed> |
==Other original publications== | ==Other original publications== | ||
Revision as of 07:56, 21 May 2014
- Description: cell shape-determining protein, forms filaments, the polymers control/restrict the mobility of the cell wall elongation enzyme complex, required for LytE activity
| Gene name | mreB |
| Synonyms | divIVB |
| Essential | yes PubMed |
| Product | cell shape-determining protein |
| Function | cell shape determination |
| Gene expression levels in SubtiExpress: mreB | |
| Interactions involving this protein in SubtInteract: MreB | |
| MW, pI | 35 kDa, 4.901 |
| Gene length, protein length | 1011 bp, 337 aa |
| Immediate neighbours | mreC, radC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 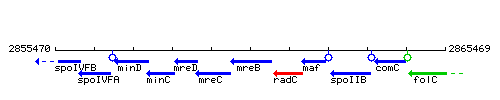
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell shape, membrane dynamics, cell envelope stress proteins (controlled by SigM, V, W, X, Y), essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28030
Phenotypes of a mutant
- essential PubMed
- the mutation can be suppressed by inactivation of ponA, ptsI, ccpA PubMed, by overexpression of YvcK PubMed, or by addition of 5 mM magnesium to the growth medium PubMed
Database entries
- BsubCyc: BSU28030
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsA/mreB family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- during logarithmic growth, MreB forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis PubMed
- forms transverse bands as cells enter the stationary phase PubMed
- forms antiparallel double filaments PubMed
- close to the inner surface of the cytoplasmic membrane PubMed
- reports on helical structures formed by MreB PubMed seem to be misinterpretation of data PubMed
- normal localization depends on the presence of glucolipids, MreB forms irregular clusters in an ugtP mutant PubMed
Database entries
- BsubCyc: BSU28030
- UniProt: Q01465
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in the labs of Jeff Errington and Boris Görke
- Antibody: available in the Jeff Errington and Peter Graumann labs
Labs working on this gene/protein
Jeff Errington, Newcastle University, UK homepage
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Reviews
Localization
Fusinita van den Ent, Thierry Izoré, Tanmay Am Bharat, Christopher M Johnson, Jan Löwe
Bacterial actin MreB forms antiparallel double filaments.
Elife: 2014, 3;e02634
[PubMed:24843005]
[WorldCat.org]
[DOI]
(I e)
Philipp V Olshausen, Hervé Joël Defeu Soufo, Kai Wicker, Rainer Heintzmann, Peter L Graumann, Alexander Rohrbach
Superresolution imaging of dynamic MreB filaments in B. subtilis--a multiple-motor-driven transport?
Biophys J: 2013, 105(5);1171-81
[PubMed:24010660]
[WorldCat.org]
[DOI]
(I p)
Christian Reimold, Herve Joel Defeu Soufo, Felix Dempwolff, Peter L Graumann
Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology.
Mol Biol Cell: 2013, 24(15);2340-9
[PubMed:23783036]
[WorldCat.org]
[DOI]
(I p)
Felix Dempwolff, Christian Reimold, Michael Reth, Peter L Graumann
Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system.
PLoS One: 2011, 6(11);e27035
[PubMed:22069484]
[WorldCat.org]
[DOI]
(I p)
Ethan C Garner, Remi Bernard, Wenqin Wang, Xiaowei Zhuang, David Z Rudner, Tim Mitchison
Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis.
Science: 2011, 333(6039);222-5
[PubMed:21636745]
[WorldCat.org]
[DOI]
(I p)
Julia Domínguez-Escobar, Arnaud Chastanet, Alvaro H Crevenna, Vincent Fromion, Roland Wedlich-Söldner, Rut Carballido-López
Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria.
Science: 2011, 333(6039);225-8
[PubMed:21636744]
[WorldCat.org]
[DOI]
(I p)
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
Hervé Joël Defeu Soufo, Peter L Graumann
Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB.
Mol Microbiol: 2006, 62(5);1340-56
[PubMed:17064365]
[WorldCat.org]
[DOI]
(P p)
Other original publications