Difference between revisions of "YvcK"
| Line 15: | Line 15: | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU34760 yvcK] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU34760 yvcK] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=YvcK YvcK] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 34 kDa, 5.408 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 34 kDa, 5.408 | ||
| Line 38: | Line 40: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[carbon core metabolism]]}}, {{SubtiWiki category|[[cell shape]]}} | + | {{SubtiWiki category|[[carbon core metabolism]]}}, {{SubtiWiki category|[[cell shape]]}}, |
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 76: | Line 79: | ||
* '''Domains:''' | * '''Domains:''' | ||
| + | ** phosphorylated on Thr-304 by [[PrkC]], dephosphorylated by [[PrpC]] {{PubMed|25012659}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| Line 84: | Line 88: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[YvcK]]-[[PrkC]] {{PubMed|25012659}} | ||
| + | ** [[YvcK]]-[[PrpC]] {{PubMed|25012659}} | ||
| − | * '''[[Localization]]:''' localized as a helical-like pattern {{PubMed|21320184}} | + | * '''[[Localization]]:''' localized as a helical-like pattern {{PubMed|25012659,21320184}} |
=== Database entries === | === Database entries === | ||
| Line 146: | Line 152: | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>9237995 16272399, 21320184 </pubmed> | + | <pubmed>9237995 16272399, 21320184 25012659</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:49, 12 July 2014
- Description: essential for growth under gluconeogenetic conditions, required for correct localization of PBP1
| Gene name | yvcK |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | localization of PBP1 |
| Gene expression levels in SubtiExpress: yvcK | |
| Interactions involving this protein in SubtInteract: YvcK | |
| MW, pI | 34 kDa, 5.408 |
| Gene length, protein length | 951 bp, 317 aa |
| Immediate neighbours | yvcL, yvcJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 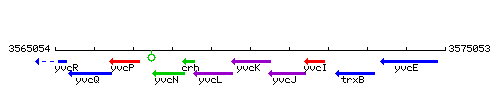
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, cell shape, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34760
Phenotypes of a mutant
- unable to grow with gluconeogenic substrates as single carbon source PubMed
- filamentous or L-shape-like aberrant morphologies (suppressed by Mg++) PubMed, this is supressed by overexpression of MreB or by deletion of ponA (PBP1 ) PubMed
Database entries
- BsubCyc: BSU34760
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: UPF0052 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: localized as a helical-like pattern PubMed
Database entries
- BsubCyc: BSU34760
- Structure:
- UniProt: O06974
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation: very weak stimuation of expression by citrate and succinate PubMed
- Regulatory mechanism:
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 96 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 246 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 555 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 282 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 459 PubMed
Biological materials
- Mutant:
- Expression vector: pGP736 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Görke lab
- Antibody:
Labs working on this gene/protein
Boris Görke, University of Göttingen, Germany Homepage
Anne Galinier, University of Marseille, France
Your additional remarks
References
Reviews
Original publications