Difference between revisions of "DltB"
(→References) |
|||
| Line 154: | Line 154: | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed> 24024634 </pubmed> | ||
| + | ==Original publications== | ||
<pubmed>14762009,12028417 ,7797557,18763711, 23980836 21856855,21926231</pubmed> | <pubmed>14762009,12028417 ,7797557,18763711, 23980836 21856855,21926231</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:25, 19 June 2014
- Description: D-alanine transfer from Dcp to undecaprenol-phosphate, alanylation of teichoic acid provides some resistance against positively charged antimicrobial peptides
| Gene name | dltB |
| Synonyms | ipa-4r |
| Essential | no |
| Product | D-alanine transfer from Dcp to undecaprenol-phosphate |
| Function | biosynthesis of teichoic acid
acid (D-alanyl transfer from Dcp to undecaprenol-phosphate) |
| Gene expression levels in SubtiExpress: dltB
acid (D-alanyl transfer from Dcp to undecaprenol-phosphate) | |
| Metabolic function and regulation of this protein in SubtiPathways: DltB | |
| MW, pI | 46 kDa, 9.944 |
| Gene length, protein length | 1185 bp, 395 aa |
| Immediate neighbours | dltA, dltC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 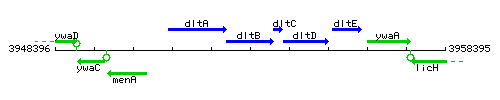
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall synthesis, transporters/ other, biosynthesis of cell wall components, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
SigD regulon, SigM regulon, SigX regulon, Spo0A regulon, stringent response, YvrHb regulon
The gene
Basic information
- Locus tag: BSU38510
Phenotypes of a mutant
- more sensitive to nisin PubMed
Database entries
- BsubCyc: BSU38510
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: membrane-bound acyltransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- membrane associated PubMed
Database entries
- BsubCyc: BSU38510
- Structure:
- UniProt: P39580
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications