Difference between revisions of "SasA"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || ppGpp synthesis independent from stringent response | |style="background:#ABCDEF;" align="center"|'''Function''' || ppGpp synthesis independent from stringent response | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38480 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38480 sasA] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 24 kDa, 6.217 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 24 kDa, 6.217 | ||
| Line 52: | Line 52: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | * a ''[[relA]] [[ | + | * a ''[[relA]] [[sasA]] [[sasB]]'' triple mutant requires branched chain amino acids, methionine and threonine for growth, the requirement can be suppressed by reduced expression of ''[[guaB]]'' or inactivation of ''[[codY]]'' {{PubMed|24163341}} |
=== Database entries === | === Database entries === | ||
| Line 70: | Line 70: | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| − | * '''Paralogous protein(s):''' [[ | + | * '''Paralogous protein(s):''' [[SasB]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 102: | Line 102: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' '' | + | * '''Operon:''' ''sasA'' {{PubMed|11866510}} |
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ywaC_3949952_3950584_-1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ywaC_3949952_3950584_-1 sasA] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' [[SigM]] {{PubMed|18179421}} [[SigW]] {{PubMed|11866510}} | * '''[[Sigma factor]]:''' [[SigM]] {{PubMed|18179421}} [[SigW]] {{PubMed|11866510}} | ||
| Line 135: | Line 135: | ||
=References= | =References= | ||
| − | <pubmed>21926231,22950019 18670626 19447912 18067544 19477419 12207695 11866510 18179421 24163341</pubmed> | + | <pubmed>21926231,22950019 18670626 19447912 18067544 19477419 12207695 11866510 18179421 24163341 24489751 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:53, 6 February 2014
- Description: (p)ppGpp synthetase, small alarmone synthetase
| Gene name | ywaC |
| Synonyms | ipa-7d |
| Essential | no |
| Product | (p)ppGpp synthetase |
| Function | ppGpp synthesis independent from stringent response |
| Gene expression levels in SubtiExpress: sasA | |
| MW, pI | 24 kDa, 6.217 |
| Gene length, protein length | 630 bp, 210 aa |
| Immediate neighbours | ywaD, menA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 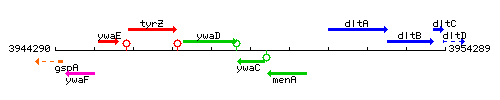
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed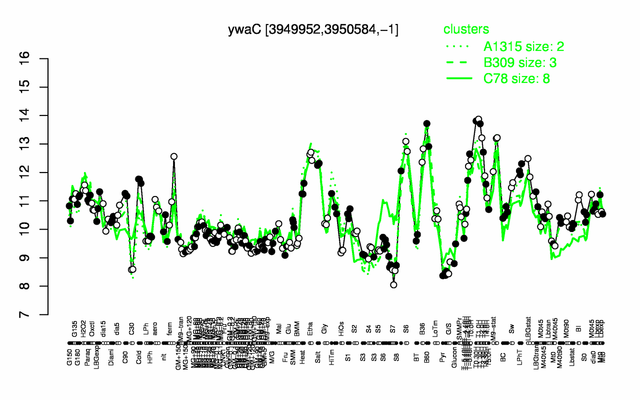
| |
Contents
Categories containing this gene/protein
metabolism of signalling nucleotides, cell envelope stress proteins (controlled by SigM, V, W, X, Y)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38480
Phenotypes of a mutant
- a relA sasA sasB triple mutant requires branched chain amino acids, methionine and threonine for growth, the requirement can be suppressed by reduced expression of guaB or inactivation of codY PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): SasB
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39583
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: sasA PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Mariangela Tabone, Virginia S Lioy, Silvia Ayora, Cristina Machón, Juan C Alonso
Role of toxin ζ and starvation responses in the sensitivity to antimicrobials.
PLoS One: 2014, 9(1);e86615
[PubMed:24489751]
[WorldCat.org]
[DOI]
(I e)
Allison Kriel, Shaun R Brinsmade, Jessica L Tse, Ashley K Tehranchi, Alycia N Bittner, Abraham L Sonenshein, Jue D Wang
GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes.
J Bacteriol: 2014, 196(1);189-201
[PubMed:24163341]
[WorldCat.org]
[DOI]
(I p)
Kazumi Tagami, Hideaki Nanamiya, Yuka Kazo, Marie Maehashi, Shota Suzuki, Eri Namba, Masahiro Hoshiya, Ryo Hanai, Yuzuru Tozawa, Takuya Morimoto, Naotake Ogasawara, Yasushi Kageyama, Katsutoshi Ara, Katsuya Ozaki, Masaki Yoshida, Haruko Kuroiwa, Tsuneyoshi Kuroiwa, Yoshiaki Ohashi, Fujio Kawamura
Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome.
Microbiologyopen: 2012, 1(2);115-34
[PubMed:22950019]
[WorldCat.org]
[DOI]
(I p)
Veronica Guariglia-Oropeza, John D Helmann
Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids.
J Bacteriol: 2011, 193(22);6223-32
[PubMed:21926231]
[WorldCat.org]
[DOI]
(I p)
Michael A D'Elia, Kathryn E Millar, Amit P Bhavsar, Ana M Tomljenovic, Bernd Hutter, Christoph Schaab, Gabriel Moreno-Hagelsieb, Eric D Brown
Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis.
Chem Biol: 2009, 16(5);548-56
[PubMed:19477419]
[WorldCat.org]
[DOI]
(I p)
Yousuke Natori, Kazumi Tagami, Kana Murakami, Sawako Yoshida, Osamu Tanigawa, Yoonsuh Moh, Kenta Masuda, Tetsuya Wada, Shota Suzuki, Hideaki Nanamiya, Yuzuru Tozawa, Fujio Kawamura
Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC.
J Bacteriol: 2009, 191(14);4555-61
[PubMed:19447912]
[WorldCat.org]
[DOI]
(I p)
Anjana Srivatsan, Yi Han, Jianlan Peng, Ashley K Tehranchi, Richard Gibbs, Jue D Wang, Rui Chen
High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies.
PLoS Genet: 2008, 4(8);e1000139
[PubMed:18670626]
[WorldCat.org]
[DOI]
(I e)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Hideaki Nanamiya, Koji Kasai, Akira Nozawa, Choong-Soo Yun, Takakuni Narisawa, Kana Murakami, Yousuke Natori, Fujio Kawamura, Yuzuru Tozawa
Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis.
Mol Microbiol: 2008, 67(2);291-304
[PubMed:18067544]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Tao Wang, Rick Ye, John D Helmann
Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons.
Mol Microbiol: 2002, 45(5);1267-76
[PubMed:12207695]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Phil A Kobel, Maud M Morshedi, Ming Fang Winston Wu, Chris Paddon, John D Helmann
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.
J Mol Biol: 2002, 316(3);443-57
[PubMed:11866510]
[WorldCat.org]
[DOI]
(P p)