Difference between revisions of "RsbR"
| Line 43: | Line 43: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[sigma factors and their control]]}}, | {{SubtiWiki category|[[sigma factors and their control]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 81: | Line 82: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
** RsbRA is composed of an N-terminal nonheme globin domain and a highly conserved C-terminal STAS (Sulphate Transporter and AntiSigma factor antagonist) domain. The C-terminal STAS domain is the target of the serine/threonine-specific kinase [[RsbT]] (see below). | ** RsbRA is composed of an N-terminal nonheme globin domain and a highly conserved C-terminal STAS (Sulphate Transporter and AntiSigma factor antagonist) domain. The C-terminal STAS domain is the target of the serine/threonine-specific kinase [[RsbT]] (see below). | ||
* '''Modification:''' phosphorylation on Thr-171 and Thr-205 by [[RsbT]] {{PubMed|21362065}} | * '''Modification:''' phosphorylation on Thr-171 and Thr-205 by [[RsbT]] {{PubMed|21362065}} | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 125: | Line 126: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 153: | Line 155: | ||
==Original Articles== | ==Original Articles== | ||
| − | + | <pubmed>8002610,8682769,8682789, 17726680,10781545,15583165, 8824586,10329124,17158665, 9179850,8808936,15312768, 11244072,15342582,15378759, 12950928, 15466036, 9179850, 8955331, 18832644 ,17726680 ,17218307 20019076 21602359 23320651,21362065,20935101,22287516,22609918 </pubmed> | |
| − | <pubmed>8002610,8682769,8682789, 17726680,10781545,15583165, 8824586,10329124,17158665, 9179850,8808936,15312768, 11244072,15342582,, 12950928, 15466036, 9179850, 8955331, 18832644 ,17726680 ,17218307 20019076 21602359 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:45, 5 March 2014
- Description: activator of RsbT kinase activity, stressosome sensor protein
| Gene name | rsbR |
| Synonyms | ycxR, rsbRA |
| Essential | no |
| Product | activator of RsbT kinase activity, stressosome sensor protein |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: rsbR | |
| Interactions involving this protein in SubtInteract: RbsR | |
| Metabolic function and regulation of this protein in SubtiPathways: rsbR | |
| MW, pI | 30 kDa, 4.731 |
| Gene length, protein length | 822 bp, 274 aa |
| Immediate neighbours | ndoA, rsbS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 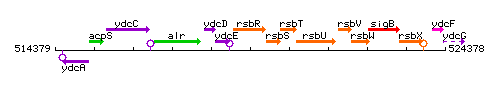
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed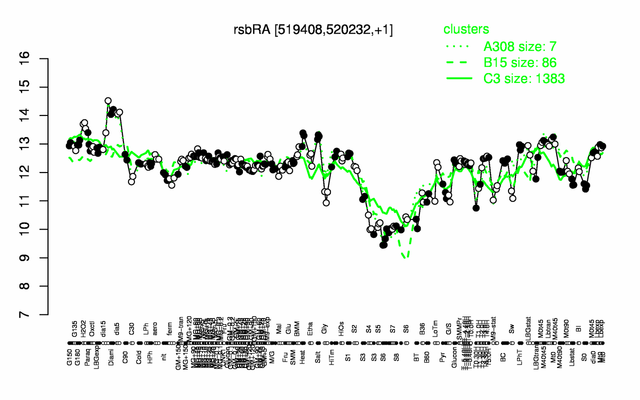
| |
Contents
Categories containing this gene/protein
sigma factors and their control, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04670
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains:
- RsbRA is composed of an N-terminal nonheme globin domain and a highly conserved C-terminal STAS (Sulphate Transporter and AntiSigma factor antagonist) domain. The C-terminal STAS domain is the target of the serine/threonine-specific kinase RsbT (see below).
- Effectors of protein activity:
- component of the stressosome
Database entries
- UniProt: P42409
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
- Rick Lewis, Newcastle, UK homepage
Your additional remarks
References
Reviews
Marcel Jurk, Philipp Schramm, Peter Schmieder
The blue-light receptor YtvA from Bacillus subtilis is permanently incorporated into the stressosome independent of the illumination state.
Biochem Biophys Res Commun: 2013, 432(3);499-503
[PubMed:23416074]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Richard J Lewis
The stressosome: molecular architecture of a signalling hub.
Biochem Soc Trans: 2010, 38(4);928-33
[PubMed:20658979]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Richard J Lewis
The Bacillus subtilis stressosome: A signal integration and transduction hub.
Commun Integr Biol: 2008, 1(2);182-4
[PubMed:19704888]
[WorldCat.org]
[DOI]
(I p)
Jan Pané-Farré, Richard J Lewis, Jörg Stülke
The RsbRST stress module in bacteria: a signalling system that may interact with different output modules.
J Mol Microbiol Biotechnol: 2005, 9(2);65-76
[PubMed:16319496]
[WorldCat.org]
[DOI]
(P p)
Original Articles
Ulf W Liebal, Thomas Millat, Jon Marles-Wright, Richard J Lewis, Olaf Wolkenhauer
Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT.
BMC Syst Biol: 2013, 7;3
[PubMed:23320651]
[WorldCat.org]
[DOI]
(I e)
Tatiana A Gaidenko, Xiaomei Bie, Enoch P Baldwin, Chester W Price
Two surfaces of a conserved interdomain linker differentially affect output from the RST sensing module of the Bacillus subtilis stressosome.
J Bacteriol: 2012, 194(15);3913-21
[PubMed:22609918]
[WorldCat.org]
[DOI]
(I p)
Jeroen B van der Steen, Marcela Avila-Pérez, Doreen Knippert, Angie Vreugdenhil, Pascal van Alphen, Klaas J Hellingwerf
Differentiation of function among the RsbR paralogs in the general stress response of Bacillus subtilis with regard to light perception.
J Bacteriol: 2012, 194(7);1708-16
[PubMed:22287516]
[WorldCat.org]
[DOI]
(I p)
Tatiana A Gaidenko, Xiaomei Bie, Enoch P Baldwin, Chester W Price
Substitutions in the presumed sensing domain of the Bacillus subtilis stressosome affect its basal output but not response to environmental signals.
J Bacteriol: 2011, 193(14);3588-97
[PubMed:21602359]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Stephan Schulz, Katrin Gronau, Dörte Becher, Michael Hecker, Chester W Price
In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB.
Mol Microbiol: 2011, 80(3);798-810
[PubMed:21362065]
[WorldCat.org]
[DOI]
(I p)
Luis Martinez, Adam Reeves, William Haldenwang
Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σ(B) activation following exposure to either physical or nutritional stress.
J Bacteriol: 2010, 192(23);6279-86
[PubMed:20935101]
[WorldCat.org]
[DOI]
(I p)
Adam Reeves, Luis Martinez, William Haldenwang
Expression of, and in vivo stressosome formation by, single members of the RsbR protein family in Bacillus subtilis.
Microbiology (Reading): 2010, 156(Pt 4);990-998
[PubMed:20019076]
[WorldCat.org]
[DOI]
(I p)
Jon Marles-Wright, Tim Grant, Olivier Delumeau, Gijs van Duinen, Susan J Firbank, Peter J Lewis, James W Murray, Joseph A Newman, Maureen B Quin, Paul R Race, Alexis Rohou, Willem Tichelaar, Marin van Heel, Richard J Lewis
Molecular architecture of the "stressosome," a signal integration and transduction hub.
Science: 2008, 322(5898);92-6
[PubMed:18832644]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Adam Reeves, W G Haldenwang
Isolation and characterization of dominant mutations in the Bacillus subtilis stressosome components RsbR and RsbS.
J Bacteriol: 2007, 189(5);1531-41
[PubMed:17158665]
[WorldCat.org]
[DOI]
(P p)
Shrin Kuo, Shuyu Zhang, Robyn L Woodbury, W G Haldenwang
Associations between Bacillus subtilis sigmaB regulators in cell extracts.
Microbiology (Reading): 2004, 150(Pt 12);4125-36
[PubMed:15583165]
[WorldCat.org]
[DOI]
(P p)
Chien-Cheng Chen, Michael D Yudkin, Olivier Delumeau
Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis.
J Bacteriol: 2004, 186(20);6830-6
[PubMed:15466036]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis.
J Bacteriol: 2004, 186(18);6124-32
[PubMed:15342582]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis.
J Mol Biol: 2004, 341(1);135-50
[PubMed:15312768]
[WorldCat.org]
[DOI]
(P p)
Chien-Cheng Chen, Richard J Lewis, Robin Harris, Michael D Yudkin, Olivier Delumeau
A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1657-69
[PubMed:12950928]
[WorldCat.org]
[DOI]
(P p)
S Zhang, J M Scott, W G Haldenwang
Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B).
J Bacteriol: 2001, 183(7);2316-21
[PubMed:11244072]
[WorldCat.org]
[DOI]
(P p)
J M Scott, J Ju, T Mitchell, W G Haldenwang
The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes.
J Bacteriol: 2000, 182(10);2771-7
[PubMed:10781545]
[WorldCat.org]
[DOI]
(P p)
T A Gaidenko, X Yang, Y M Lee, C W Price
Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis.
J Mol Biol: 1999, 288(1);29-39
[PubMed:10329124]
[WorldCat.org]
[DOI]
(P p)
S Akbar, C M Kang, T A Gaidenko, C W Price
Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis.
Mol Microbiol: 1997, 24(3);567-78
[PubMed:9179850]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
The yeast two-hybrid system detects interactions between Bacillus subtilis sigmaB regulators.
J Bacteriol: 1996, 178(23);7020-3
[PubMed:8955331]
[WorldCat.org]
[DOI]
(P p)
X Yang, C M Kang, M S Brody, C W Price
Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor.
Genes Dev: 1996, 10(18);2265-75
[PubMed:8824586]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, W G Haldenwang
Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities.
J Bacteriol: 1996, 178(18);5456-63
[PubMed:8808936]
[WorldCat.org]
[DOI]
(P p)
C M Kang, M S Brody, S Akbar, X Yang, C W Price
Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress.
J Bacteriol: 1996, 178(13);3846-53
[PubMed:8682789]
[WorldCat.org]
[DOI]
(P p)
A Dufour, U Voelker, A Voelker, W G Haldenwang
Relative levels and fractionation properties of Bacillus subtilis σ(B) and its regulators during balanced growth and stress.
J Bacteriol: 1996, 178(13);3701-9 sigma
[PubMed:8682769]
[WorldCat.org]
[DOI]
(P p)
A A Wise, C W Price
Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals.
J Bacteriol: 1995, 177(1);123-33
[PubMed:8002610]
[WorldCat.org]
[DOI]
(P p)