Difference between revisions of "YtvA"
(→Original publications) |
|||
| Line 146: | Line 146: | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed> 19581299 16923906,17575448, 17764689, 17443319 17200735 16923909 16797012 16022561 15339223 11964249 11157946 19891470 20062844 19783626 20800068 22247770 23047813 23416074 23456923 23817467 21259411,21388385, 21410163, 21851109,22287516 23861663 23828427 23900620 23940057 </pubmed> | + | <pubmed> 19581299 16923906,17575448, 17764689, 17443319 17200735 16923909 16797012 16022561 15339223 11964249 11157946 19891470 20062844 19783626 20800068 22247770 23047813 23416074 23456923 23817467 21259411,21388385, 21410163, 21851109,22287516 23861663 23828427 23900620 23940057 24171435</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:22, 12 November 2013
- Description: blue light sensor, positive regulation of SigB activity under conditions of blue light
| Gene name | ytvA |
| Synonyms | |
| Essential | no |
| Product | blue light receptor, flavoprotein |
| Function | control of SigB activity |
| Gene expression levels in SubtiExpress: ytvA | |
| Interactions involving this protein in SubtInteract: YtvA | |
| MW, pI | 29 kDa, 4.69 |
| Gene length, protein length | 783 bp, 261 aa |
| Immediate neighbours | ytvB, yttB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 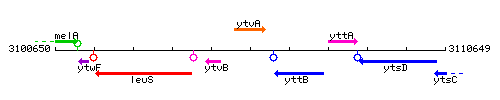
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed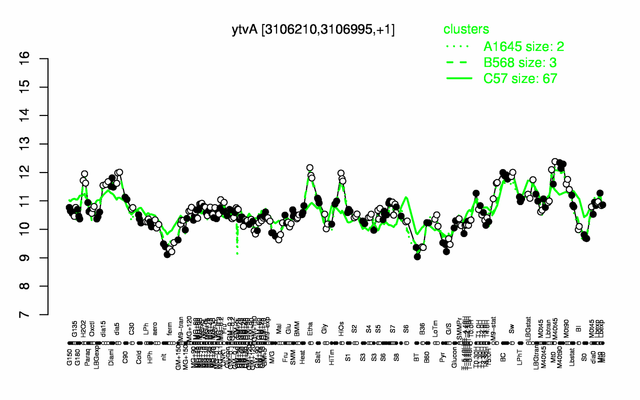
| |
Contents
Categories containing this gene/protein
sigma factors and their control
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30340
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains: LOV domain, STAS domain
- Modification:
- Cofactor(s): FMN
- Effectors of protein activity:
Database entries
- UniProt: O34627
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews
Original publications