Difference between revisions of "RpsL"
| Line 60: | Line 60: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01100&redirect=T BSU01100] | ||
* '''DBTBS entry:''' no entry | * '''DBTBS entry:''' no entry | ||
| Line 99: | Line 100: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU01100&redirect=T BSU01100] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 12:49, 2 April 2014
- Description: ribosomal protein
| Gene name | rpsL |
| Synonyms | strA, fun |
| Essential | yes PubMed |
| Product | ribosomal protein S12 (BS12) |
| Function | translation |
| Gene expression levels in SubtiExpress: rpsL | |
| Interactions involving this protein in SubtInteract: RpsL | |
| MW, pI | 15 kDa, 11.721 |
| Gene length, protein length | 414 bp, 138 aa |
| Immediate neighbours | ybxF, rpsG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed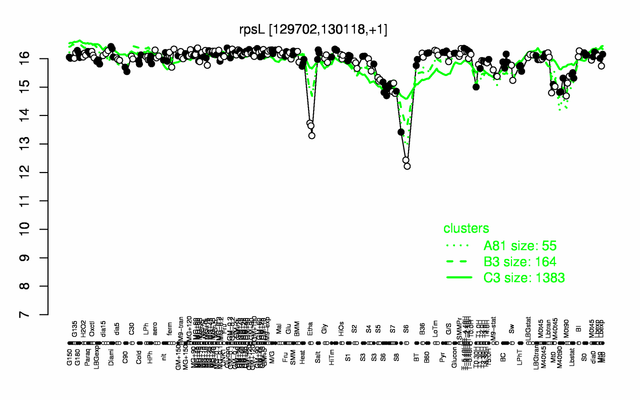
| |
Contents
Categories containing this gene/protein
translation, essential genes, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01100
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU01100
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ribosomal protein S12P family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-123 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU01100
- Structure:
- UniProt: P21472
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yun Chen, Shu Feng, Veerendra Kumar, Rya Ero, Yong-Gui Gao
Structure of EF-G-ribosome complex in a pretranslocation state.
Nat Struct Mol Biol: 2013, 20(9);1077-84
[PubMed:23912278]
[WorldCat.org]
[DOI]
(I p)
Genki Akanuma, Hideaki Nanamiya, Yousuke Natori, Koichi Yano, Shota Suzuki, Shuya Omata, Morio Ishizuka, Yasuhiko Sekine, Fujio Kawamura
Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation.
J Bacteriol: 2012, 194(22);6282-91
[PubMed:23002217]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Matthew A Lauber, William E Running, James P Reilly
B. subtilis ribosomal proteins: structural homology and post-translational modifications.
J Proteome Res: 2009, 8(9);4193-206
[PubMed:19653700]
[WorldCat.org]
[DOI]
(P p)
Kazuhiko Kurosawa, Takeshi Hosaka, Norimasa Tamehiro, Takashi Inaoka, Kozo Ochi
Improvement of alpha-amylase production by modulation of ribosomal component protein S12 in Bacillus subtilis 168.
Appl Environ Microbiol: 2006, 72(1);71-7
[PubMed:16391027]
[WorldCat.org]
[DOI]
(P p)
T Inaoka, K Kasai, K Ochi
Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis.
J Bacteriol: 2001, 183(17);4958-63
[PubMed:11489846]
[WorldCat.org]
[DOI]
(P p)