Difference between revisions of "Lip"
Raphael2215 (talk | contribs) (→Database entries) |
(→References) |
||
| Line 138: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>22996591 22267088 23404771 21477088,22246996 23639749 21815947 15812018,12523966,18721749,12218047,16342303, 12951259 11583117 11491291 18383241,19180538, 8396026, 19883129 18840696 18053819</pubmed> | + | <pubmed>22996591 22267088 23404771 21477088,22246996 23639749 21815947 24402332 15812018,12523966,18721749,12218047,16342303, 12951259 11583117 11491291 18383241,19180538, 8396026, 19883129 18840696 18053819</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:36, 10 January 2014
- Description: extracellular lipase
| Gene name | lip |
| Synonyms | lipA |
| Essential | no |
| Product | extracellular lipase |
| Function | lipid degradation |
| Gene expression levels in SubtiExpress: lip | |
| MW, pI | 22 kDa, 10.059 |
| Gene length, protein length | 636 bp, 212 aa |
| Immediate neighbours | ansZ, yczC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 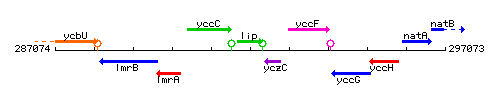
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed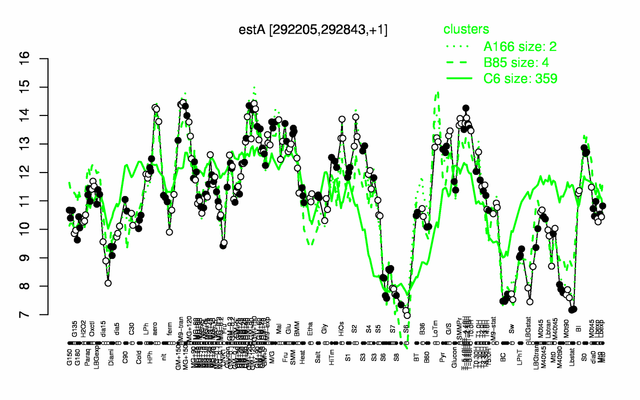
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02700
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): LipB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P37957
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Virender Kumar, Poornima Yedavalli, Vishal Gupta, Nalam Madhusudhana Rao
Engineering lipase A from mesophilic Bacillus subtilis for activity at low temperatures.
Protein Eng Des Sel: 2014, 27(3);73-82
[PubMed:24402332]
[WorldCat.org]
[DOI]
(I p)
Md Zahid Kamal, Jamshaid Ali, Nalam Madhusudhana Rao
Binding of bis-ANS to Bacillus subtilis lipase: a combined computational and experimental investigation.
Biochim Biophys Acta: 2013, 1834(8);1501-9
[PubMed:23639749]
[WorldCat.org]
[DOI]
(P p)
Poornima Yedavalli, Nalam Madhusudhana Rao
Engineering the loops in a lipase for stability in DMSO.
Protein Eng Des Sel: 2013, 26(4);317-24
[PubMed:23404771]
[WorldCat.org]
[DOI]
(I p)
Wojciech Augustyniak, Hans Wienk, Rolf Boelens, Manfred T Reetz
¹H, ¹³C and ¹⁵N resonance assignments of wild-type Bacillus subtilis Lipase A and its mutant evolved towards thermostability.
Biomol NMR Assign: 2013, 7(2);249-52
[PubMed:22996591]
[WorldCat.org]
[DOI]
(I p)
Wojciech Augustyniak, Agnieszka A Brzezinska, Tjaard Pijning, Hans Wienk, Rolf Boelens, Bauke W Dijkstra, Manfred T Reetz
Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: factors contributing to increased activity retention.
Protein Sci: 2012, 21(4);487-97
[PubMed:22267088]
[WorldCat.org]
[DOI]
(I p)
Shoeb Ahmad, Virender Kumar, K Bhanu Ramanand, N Madhusudhana Rao
Probing protein stability and proteolytic resistance by loop scanning: a comprehensive mutational analysis.
Protein Sci: 2012, 21(3);433-46
[PubMed:22246996]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Zhong Ni, Peng Zhou, Xin Jin, Xian-Fu Lin
Integrating In Silico and In vitro approaches to dissect the stereoselectivity of Bacillus subtilis lipase A toward ketoprofen vinyl ester.
Chem Biol Drug Des: 2011, 78(2);301-8
[PubMed:21477088]
[WorldCat.org]
[DOI]
(I p)
Bo Chen, Zhen Cai, Wei Wu, Yunlong Huang, Juergen Pleiss, Zhanglin Lin
Morphing activity between structurally similar enzymes: from heme-free bromoperoxidase to lipase.
Biochemistry: 2009, 48(48);11496-504
[PubMed:19883129]
[WorldCat.org]
[DOI]
(I p)
Thijs R H M Kouwen, René van der Ploeg, Haike Antelmann, Michael Hecker, Georg Homuth, Ulrike Mäder, Jan Maarten van Dijl
Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics.
Proteomics: 2009, 9(4);1018-32
[PubMed:19180538]
[WorldCat.org]
[DOI]
(I p)
Allison V Banse, Arnaud Chastanet, Lilah Rahn-Lee, Errett C Hobbs, Richard Losick
Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2008, 105(40);15547-52
[PubMed:18840696]
[WorldCat.org]
[DOI]
(I p)
Ykelien L Boersma, Tjaard Pijning, Margriet S Bosma, Almer M van der Sloot, Luís F Godinho, Melloney J Dröge, Remko T Winter, Gertie van Pouderoyen, Bauke W Dijkstra, Wim J Quax
Loop grafting of Bacillus subtilis lipase A: inversion of enantioselectivity.
Chem Biol: 2008, 15(8);782-9
[PubMed:18721749]
[WorldCat.org]
[DOI]
(P p)
Ykelien L Boersma, Melloney J Dröge, Almer M van der Sloot, Tjaard Pijning, Robbert H Cool, Bauke W Dijkstra, Wim J Quax
A novel genetic selection system for improved enantioselectivity of Bacillus subtilis lipase A.
Chembiochem: 2008, 9(7);1110-5
[PubMed:18383241]
[WorldCat.org]
[DOI]
(I p)
Eerappa Rajakumara, Priyamvada Acharya, Shoeb Ahmad, Rajan Sankaranaryanan, Nalam M Rao
Structural basis for the remarkable stability of Bacillus subtilis lipase (Lip A) at low pH.
Biochim Biophys Acta: 2008, 1784(2);302-11
[PubMed:18053819]
[WorldCat.org]
[DOI]
(P p)
Melloney J Dröge, Ykelien L Boersma, Gertie van Pouderoyen, Titia E Vrenken, Carsten J Rüggeberg, Manfred T Reetz, Bauke W Dijkstra, Wim J Quax
Directed evolution of Bacillus subtilis lipase A by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection.
Chembiochem: 2006, 7(1);149-57
[PubMed:16342303]
[WorldCat.org]
[DOI]
(P p)
Helga Westers, Peter G Braun, Lidia Westers, Haike Antelmann, Michael Hecker, Jan D H Jongbloed, Hirofumi Yoshikawa, Teruo Tanaka, Jan Maarten van Dijl, Wim J Quax
Genes involved in SkfA killing factor production protect a Bacillus subtilis lipase against proteolysis.
Appl Environ Microbiol: 2005, 71(4);1899-908
[PubMed:15812018]
[WorldCat.org]
[DOI]
(P p)
Thorsten Eggert, Ulf Brockmeier, Melloney J Dröge, Wim J Quax, Karl-Erich Jaeger
Extracellular lipases from Bacillus subtilis: regulation of gene expression and enzyme activity by amino acid supply and external pH.
FEMS Microbiol Lett: 2003, 225(2);319-24
[PubMed:12951259]
[WorldCat.org]
[DOI]
(P p)
Melloney J Dröge, Carsten J Rüggeberg, Almer M van der Sloot, Judith Schimmel, Dolf Swaving Dijkstra, Raymond M D Verhaert, Manfred T Reetz, Wim J Quax
Binding of phage displayed Bacillus subtilis lipase A to a phosphonate suicide inhibitor.
J Biotechnol: 2003, 101(1);19-28
[PubMed:12523966]
[WorldCat.org]
[DOI]
(P p)
Jan D H Jongbloed, Haike Antelmann, Michael Hecker, Reindert Nijland, Sierd Bron, Ulla Airaksinen, Frens Pries, Wim J Quax, Jan Maarten van Dijl, Peter G Braun
Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis.
J Biol Chem: 2002, 277(46);44068-78
[PubMed:12218047]
[WorldCat.org]
[DOI]
(P p)
T Eggert, G van Pouderoyen, B W Dijkstra, K E Jaeger
Lipolytic enzymes LipA and LipB from Bacillus subtilis differ in regulation of gene expression, biochemical properties, and three-dimensional structure.
FEBS Lett: 2001, 502(3);89-92
[PubMed:11583117]
[WorldCat.org]
[DOI]
(P p)
G van Pouderoyen, T Eggert, K E Jaeger, B W Dijkstra
The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme.
J Mol Biol: 2001, 309(1);215-26
[PubMed:11491291]
[WorldCat.org]
[DOI]
(P p)
E Lesuisse, K Schanck, C Colson
Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme.
Eur J Biochem: 1993, 216(1);155-60
[PubMed:8396026]
[WorldCat.org]
[DOI]
(P p)