Difference between revisions of "Csn"
(→References) |
|||
| Line 135: | Line 135: | ||
=References= | =References= | ||
| − | <pubmed>11065371,18957862, 22031025 20817675</pubmed> | + | <pubmed>11065371,18957862, 22031025 20817675 12401130 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:47, 8 July 2013
- Description: chitosanase
| Gene name | csn |
| Synonyms | |
| Essential | no |
| Product | chitosanase |
| Function | chitin degradation |
| Gene expression levels in SubtiExpress: csn | |
| MW, pI | 31 kDa, 8.892 |
| Gene length, protein length | 831 bp, 277 aa |
| Immediate neighbours | yraM, yraL |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 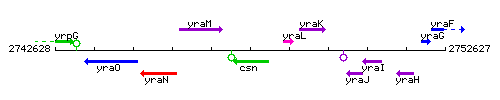
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed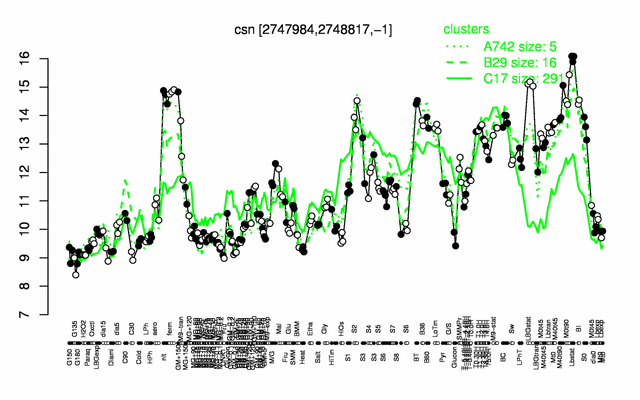
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU26890
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endohydrolysis of beta-(1->4)-linkages between D-glucosamine residues in a partly acetylated chitosan (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 46 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- extracellular (signal peptide), major constituent of the secretome PubMed
Database entries
- Structure:
- UniProt: O07921
- KEGG entry: [2]
- E.C. number: 3.2.1.132
Additional information
Expression and regulation
- Operon:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Chulhong Oh, Mahanama De Zoysa, Do-Hyung Kang, Youngdeuk Lee, Ilson Whang, Chamilani Nikapitiya, Soo-Jin Heo, Kon-Tak Yoon, Abu Affan, Jehee Lee
Isolation, purification, and enzymatic characterization of extracellular chitosanase from marine bacterium Bacillus subtilis CH2.
J Microbiol Biotechnol: 2011, 21(10);1021-5
[PubMed:22031025]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Anne Colomer-Pallas, Yannick Pereira, Marie-Françoise Petit-Glatron, Régis Chambert
Calcium triggers the refolding of Bacillus subtilis chitosanase.
Biochem J: 2003, 369(Pt 3);731-8
[PubMed:12401130]
[WorldCat.org]
[DOI]
(P p)
Luis A Rivas, Vı Ctor Parro, Mercedes Moreno-Paz, Rafael P Mellado
The Bacillus subtilis 168 csn gene encodes a chitosanase with similar properties to a streptomyces enzyme.
Microbiology (Reading): 2000, 146 ( Pt 11);2929-2936
[PubMed:11065371]
[WorldCat.org]
[DOI]
(P p)