Difference between revisions of "PGP380"
| Line 8: | Line 8: | ||
*'''M13_puc_for:''' 5‘-GTAAAACGACGGCCAGTG-3‘ | *'''M13_puc_for:''' 5‘-GTAAAACGACGGCCAGTG-3‘ | ||
*'''M13_puc_rev:''' 5‘-GGAAACAGCTATGACCATG-3‘ | *'''M13_puc_rev:''' 5‘-GGAAACAGCTATGACCATG-3‘ | ||
| + | *'''FX125 (rev; primes -73bp of MCS):''' 5‘-GGCTCGTATGTTGTGTGG-3‘ | ||
| + | *'''JN54 (fwd; primes +148bp of MCS):''' 5‘-GTGAAAAATGAGCCGAAAGCAG-3‘ | ||
Latest revision as of 08:13, 29 July 2014
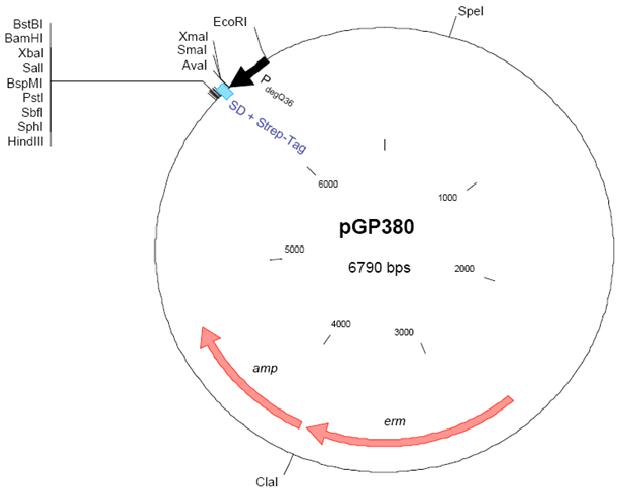
Application:
The vector was constructed in the Stülke lab and it is suitable for constitutive overexpression of N-terminally Strep-tagged proteins in B. subtilis. The plasmid confers resistance to ampicillin and erythromycin in E. coli and B. subtilis, respectively. pGP380 can be used for the SPINE method. pGP382 is similar to the vector pGP380 which is derived from vectors pBQ200 and pHT315.
Sequencing primers:
- M13_puc_for: 5‘-GTAAAACGACGGCCAGTG-3‘
- M13_puc_rev: 5‘-GGAAACAGCTATGACCATG-3‘
- FX125 (rev; primes -73bp of MCS): 5‘-GGCTCGTATGTTGTGTGG-3‘
- JN54 (fwd; primes +148bp of MCS): 5‘-GTGAAAAATGAGCCGAAAGCAG-3‘