Difference between revisions of "FatR"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[electron transport/ other]]}}, | {{SubtiWiki category|[[electron transport/ other]]}}, | ||
| − | + | {{SubtiWiki category|[[lipid metabolism/ other]]}}, | |
{{SubtiWiki category|[[transcription factors and their control]]}}, | {{SubtiWiki category|[[transcription factors and their control]]}}, | ||
| − | {{SubtiWiki category|[[cell envelope stress proteins (controlled by SigM, V, W, X, Y)]]}} | + | {{SubtiWiki category|[[cell envelope stress proteins (controlled by SigM, V, W, X, Y)]]}}, |
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 88: | Line 85: | ||
* '''Modification:''' | * '''Modification:''' | ||
| + | ** phosphorylated by [[PtkA]] on Tyr-45, this results in displacement of FatR from the DNA {{PubMed|23939619}} | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 94: | Line 92: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** [[FatR]]-[[TkmA]] {{PubMed|23939619}} | ||
| + | ** [[FatR]]-[[PtkA]] {{PubMed|23939619}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 147: | Line 147: | ||
=References= | =References= | ||
| − | + | <pubmed>11734890,11574077,11377867,10917605,12207695,9636707 23939619 21926231</pubmed> | |
| − | <pubmed>11734890,11574077,11377867,10917605,12207695,9636707</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:23, 14 August 2013
| Gene name | fatR |
| Synonyms | yrhI, bscR |
| Essential | no |
| Product | transcriptional repressor of the fatR-yrhJ operon (TetR family) |
| Function | unknown |
| Gene expression levels in SubtiExpress: fatR | |
| MW, pI | 22 kDa, 5.621 |
| Gene length, protein length | 582 bp, 194 aa |
| Immediate neighbours | yrhJ, yrhH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 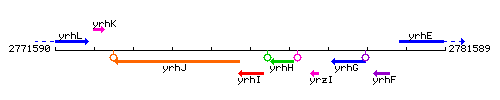
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed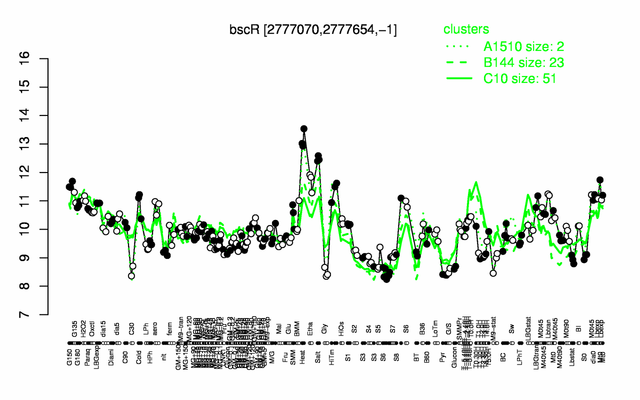
| |
Contents
Categories containing this gene/protein
electron transport/ other, lipid metabolism/ other, transcription factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), phosphoproteins
This gene is a member of the following regulons
FatR regulon, SigM regulon, SigW regulon, SigX regulon
The FatR regulon:
The gene
Basic information
- Locus tag: BSU27170
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O08335
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Abderahmane Derouiche, Vladimir Bidnenko, Rosa Grenha, Nathalie Pigonneau, Magali Ventroux, Mirita Franz-Wachtel, Sylvie Nessler, Marie-Françoise Noirot-Gros, Ivan Mijakovic
Interaction of bacterial fatty-acid-displaced regulators with DNA is interrupted by tyrosine phosphorylation in the helix-turn-helix domain.
Nucleic Acids Res: 2013, 41(20);9371-81
[PubMed:23939619]
[WorldCat.org]
[DOI]
(I p)
Veronica Guariglia-Oropeza, John D Helmann
Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids.
J Bacteriol: 2011, 193(22);6223-32
[PubMed:21926231]
[WorldCat.org]
[DOI]
(I p)
Min Cao, Tao Wang, Rick Ye, John D Helmann
Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons.
Mol Microbiol: 2002, 45(5);1267-76
[PubMed:12207695]
[WorldCat.org]
[DOI]
(P p)
M C Gustafsson, C N Palmer, C R Wolf, C von Wachenfeldt
Fatty-acid-displaced transcriptional repressor, a conserved regulator of cytochrome P450 102 transcription in Bacillus species.
Arch Microbiol: 2001, 176(6);459-64
[PubMed:11734890]
[WorldCat.org]
[DOI]
(P p)
T R Lee, H P Hsu, G C Shaw
Transcriptional regulation of the Bacillus subtilis bscR-CYP102A3 operon by the BscR repressor and differential induction of cytochrome CYP102A3 expression by oleic acid and palmitate.
J Biochem: 2001, 130(4);569-74
[PubMed:11574077]
[WorldCat.org]
[DOI]
(P p)
M C Gustafsson, C von Wachenfeldt
A novel diffusible substance can overcome the apparent AbrB repression of the Bacillus subtilis fatR promoter.
FEMS Microbiol Lett: 2001, 199(2);197-202
[PubMed:11377867]
[WorldCat.org]
[DOI]
(P p)
C N Palmer, M C Gustafsson, H Dobson, C von Wachenfeldt, C R Wolf
Adaptive responses to fatty acids are mediated by the regulated expression of cytochromes P450.
Biochem Soc Trans: 1999, 27(4);374-8
[PubMed:10917605]
[WorldCat.org]
[DOI]
(P p)
X Huang, J D Helmann
Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search.
J Mol Biol: 1998, 279(1);165-73
[PubMed:9636707]
[WorldCat.org]
[DOI]
(P p)