Difference between revisions of "FtsW"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 117: | Line 113: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ftsW_1552899_1554110_1 ftsW] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ftsW_1552899_1554110_1 ftsW] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** constitutively expressed {{PubMed|23701187}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 145: | Line 142: | ||
=References= | =References= | ||
| − | <pubmed>,17981970, 19429628 17526699 21564336</pubmed> | + | <pubmed>,17981970, 19429628 17526699 21564336 23701187</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:00, 27 May 2013
- Description: cell-division protein
| Gene name | ftsW |
| Synonyms | ylaO |
| Essential | yes PubMed |
| Product | cell-division protein |
| Function | cell division |
| Gene expression levels in SubtiExpress: ftsW | |
| Interactions involving this protein in SubtInteract: FtsW | |
| MW, pI | 43 kDa, 9.722 |
| Gene length, protein length | 1209 bp, 403 aa |
| Immediate neighbours | ylaN, pycA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 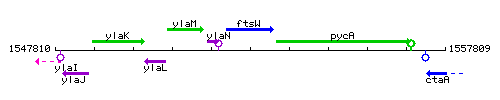
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed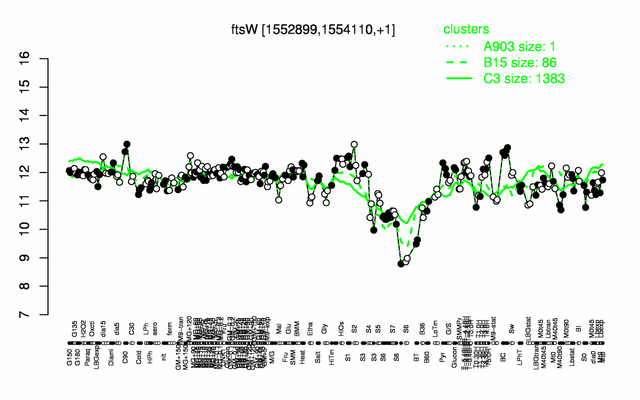
| |
Contents
Categories containing this gene/protein
cell division, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14850
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsW/rodA/spoVE family (according to Swiss-Prot) SEDS proteins (shape, elongation, division, and sporulation)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane
Database entries
- Structure:
- UniProt: O07639
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- constitutively expressed PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Erik Nico Trip, Jan-Willem Veening, Eric J Stewart, Jeff Errington, Dirk-Jan Scheffers
Balanced transcription of cell division genes in Bacillus subtilis as revealed by single cell analysis.
Environ Microbiol: 2013, 15(12);3196-209
[PubMed:23701187]
[WorldCat.org]
[DOI]
(I p)
Kenneth Briley, Peter Prepiak, Miguel J Dias, Jeanette Hahn, David Dubnau
Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis.
Mol Microbiol: 2011, 81(1);23-39
[PubMed:21564336]
[WorldCat.org]
[DOI]
(I p)
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
Gonçalo Real, Allison Fay, Avigdor Eldar, Sérgio M Pinto, Adriano O Henriques, Jonathan Dworkin
Determinants for the subcellular localization and function of a nonessential SEDS protein.
J Bacteriol: 2008, 190(1);363-76
[PubMed:17981970]
[WorldCat.org]
[DOI]
(I p)
Zuolei Lu, Michio Takeuchi, Tsutomu Sato
The LysR-type transcriptional regulator YofA controls cell division through the regulation of expression of ftsW in Bacillus subtilis.
J Bacteriol: 2007, 189(15);5642-51
[PubMed:17526699]
[WorldCat.org]
[DOI]
(P p)