Difference between revisions of "SrfAD"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 115: | Line 111: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=srfAD_402388_403116_1 srfAD] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=srfAD_402388_403116_1 srfAD] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|1715856}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|1715856}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 153: | Line 149: | ||
<pubmed>20735481 </pubmed> | <pubmed>20735481 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | + | <pubmed>18704089,16166527,17227471, 17190806,8830686, 1715856 12642660 16091051 22511326,20817675</pubmed> | |
| − | <pubmed>18704089,16166527,17227471, 17190806,8830686, 1715856 12642660 16091051 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:27, 18 June 2013
- Description: surfactin synthetase / competence
| Gene name | srfAD |
| Synonyms | comL |
| Essential | no |
| Product | surfactin synthetase / competence |
| Function | antibiotic synthesis |
| Gene expression levels in SubtiExpress: srfAD | |
| MW, pI | 27 kDa, 5.22 |
| Gene length, protein length | 726 bp, 242 aa |
| Immediate neighbours | srfAC, ycxA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 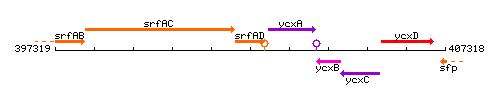
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed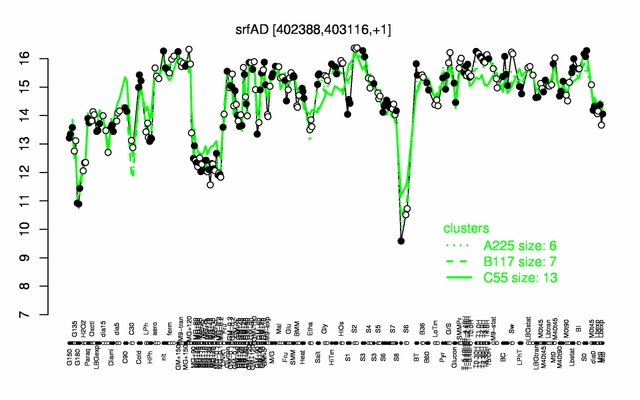
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
Abh regulon, CodY regulon, ComA regulon, PerR regulon, Spx regulon
The gene
Basic information
- Locus tag: BSU03520
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: thioesterase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: Q08788
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Massimiliano Marvasi, Pieter T Visscher, Lilliam Casillas Martinez
Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis.
FEMS Microbiol Lett: 2010, 313(1);1-9
[PubMed:20735481]
[WorldCat.org]
[DOI]
(I p)
Original publications
Ju Jung, Kyung Ok Yu, Ahmad Bazli Ramzi, Se Hoon Choe, Seung Wook Kim, Sung Ok Han
Improvement of surfactin production in Bacillus subtilis using synthetic wastewater by overexpression of specific extracellular signaling peptides, comX and phrC.
Biotechnol Bioeng: 2012, 109(9);2349-56
[PubMed:22511326]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Alexander Koglin, Frank Löhr, Frank Bernhard, Vladimir V Rogov, Dominique P Frueh, Eric R Strieter, Mohammad R Mofid, Peter Güntert, Gerhard Wagner, Christopher T Walsh, Mohamed A Marahiel, Volker Dötsch
Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature: 2008, 454(7206);907-11
[PubMed:18704089]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Yasutaro Fujita
Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression.
FEMS Microbiol Lett: 2007, 268(1);73-80
[PubMed:17227471]
[WorldCat.org]
[DOI]
(P p)
Paul D Straight, Michael A Fischbach, Christopher T Walsh, David Z Rudner, Roberto Kolter
A singular enzymatic megacomplex from Bacillus subtilis.
Proc Natl Acad Sci U S A: 2007, 104(1);305-10
[PubMed:17190806]
[WorldCat.org]
[DOI]
(P p)
Kentaro Hayashi, Taku Ohsawa, Kazuo Kobayashi, Naotake Ogasawara, Mitsuo Ogura
The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis.
J Bacteriol: 2005, 187(19);6659-67
[PubMed:16166527]
[WorldCat.org]
[DOI]
(P p)
Natalia Comella, Alan D Grossman
Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis.
Mol Microbiol: 2005, 57(4);1159-74
[PubMed:16091051]
[WorldCat.org]
[DOI]
(P p)
Shunji Nakano, Michiko M Nakano, Ying Zhang, Montira Leelakriangsak, Peter Zuber
A regulatory protein that interferes with activator-stimulated transcription in bacteria.
Proc Natl Acad Sci U S A: 2003, 100(7);4233-8
[PubMed:12642660]
[WorldCat.org]
[DOI]
(P p)
P Serror, A L Sonenshein
CodY is required for nutritional repression of Bacillus subtilis genetic competence.
J Bacteriol: 1996, 178(20);5910-5
[PubMed:8830686]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, L A Xia, P Zuber
Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis.
J Bacteriol: 1991, 173(17);5487-93
[PubMed:1715856]
[WorldCat.org]
[DOI]
(P p)