Difference between revisions of "SigX"
| Line 31: | Line 31: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=sigX_2414627_2415211_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:sigX_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=sigX_2414627_2415211_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:sigX_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU23100]] |
|- | |- | ||
|} | |} | ||
Revision as of 13:47, 16 May 2013
- Description: RNA polymerase ECF-type sigma factor SigX
| Gene name | sigX |
| Synonyms | ypuM |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigX |
| Function | cell surface properties |
| Gene expression levels in SubtiExpress: sigX | |
| Interactions involving this protein in SubtInteract: SigX | |
| MW, pI | 23 kDa, 6.086 |
| Gene length, protein length | 582 bp, 194 aa |
| Immediate neighbours | rsiX, resE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 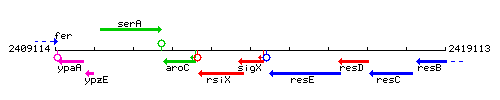
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
The SigX regulon
The gene
Basic information
- Locus tag: BSU23100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P35165
- KEGG entry: [3]
- E.C. number:
Additional information
- Expression of the SigX regulon in increased in ugtP mutants PubMed
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
The SigX regulon
Thorsten Mascher, Anna-Barbara Hachmann, John D Helmann
Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors.
J Bacteriol: 2007, 189(19);6919-27
[PubMed:17675383]
[WorldCat.org]
[DOI]
(P p)
M S Turner, J D Helmann
Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis.
J Bacteriol: 2000, 182(18);5202-10
[PubMed:10960106]
[WorldCat.org]
[DOI]
(P p)
X Huang, K L Fredrick, J D Helmann
Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon.
J Bacteriol: 1998, 180(15);3765-70
[PubMed:9683469]
[WorldCat.org]
[DOI]
(P p)
Other original publications
Michihiro Hashimoto, Takahiro Seki, Satoshi Matsuoka, Hiroshi Hara, Kei Asai, Yoshito Sadaie, Kouji Matsumoto
Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis.
Microbiology (Reading): 2013, 159(Pt 1);23-35
[PubMed:23103977]
[WorldCat.org]
[DOI]
(I p)
Satoshi Matsuoka, Minako Chiba, Yu Tanimura, Michihiro Hashimoto, Hiroshi Hara, Kouji Matsumoto
Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids.
Genes Genet Syst: 2011, 86(5);295-304
[PubMed:22362028]
[WorldCat.org]
[DOI]
(I p)
Ewan J Murray, Mark A Strauch, Nicola R Stanley-Wall
SigmaX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh.
J Bacteriol: 2009, 191(22);6822-32
[PubMed:19767430]
[WorldCat.org]
[DOI]
(I p)
Yun Luo, John D Helmann
Extracytoplasmic function sigma factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis.
J Bacteriol: 2009, 191(15);4951-8
[PubMed:19465659]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA.
J Bacteriol: 2009, 191(3);1101-5
[PubMed:19047346]
[WorldCat.org]
[DOI]
(I p)
Masakuni Serizawa, Keisuke Kodama, Hiroki Yamamoto, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses.
Biosci Biotechnol Biochem: 2005, 69(11);2155-69
[PubMed:16306698]
[WorldCat.org]
[DOI]
(P p)
Mika Yoshimura, Kei Asai, Yoshito Sadaie, Hirofumi Yoshikawa
Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors.
Microbiology (Reading): 2004, 150(Pt 3);591-599
[PubMed:14993308]
[WorldCat.org]
[DOI]
(P p)
Min Cao, John D Helmann
The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides.
J Bacteriol: 2004, 186(4);1136-46
[PubMed:14762009]
[WorldCat.org]
[DOI]
(P p)
Thorsten Mascher, Neil G Margulis, Tao Wang, Rick W Ye, John D Helmann
Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon.
Mol Microbiol: 2003, 50(5);1591-604
[PubMed:14651641]
[WorldCat.org]
[DOI]
(P p)
Min Cao, John D Helmann
Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors.
J Bacteriol: 2002, 184(22);6123-9
[PubMed:12399481]
[WorldCat.org]
[DOI]
(P p)
J Qiu, J D Helmann
The -10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function sigma factors sigma(X) and sigma(W).
J Bacteriol: 2001, 183(6);1921-7
[PubMed:11222589]
[WorldCat.org]
[DOI]
(P p)
X Huang, J D Helmann
Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search.
J Mol Biol: 1998, 279(1);165-73
[PubMed:9636707]
[WorldCat.org]
[DOI]
(P p)
S Brutsche, V Braun
SigX of Bacillus subtilis replaces the ECF sigma factor fecI of Escherichia coli and is inhibited by RsiX.
Mol Gen Genet: 1997, 256(4);416-25
[PubMed:9393439]
[WorldCat.org]
[DOI]
(P p)
X Huang, A Decatur, A Sorokin, J D Helmann
The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature.
J Bacteriol: 1997, 179(9);2915-21
[PubMed:9139908]
[WorldCat.org]
[DOI]
(P p)
V Azevedo, A Sorokin, S D Ehrlich, P Serror
The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci.
Mol Microbiol: 1993, 10(2);397-405
[PubMed:7934830]
[WorldCat.org]
[DOI]
(P p)