Difference between revisions of "PgdS"
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywtE]]'', ''[[capE]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywtE]]'', ''[[capE]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU35860 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU35860 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU35860 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU35860 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU35860 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU35860 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ywtD_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ywtD_context.gif]] | ||
Revision as of 11:26, 14 May 2013
- Description: gamma-DL-glutamyl hydrolase
| Gene name | pgdS |
| Synonyms | ywtD |
| Essential | no |
| Product | gamma-DL-glutamyl hydrolase |
| Function | polyglutamic acid degradation |
| Gene expression levels in SubtiExpress: pgdS | |
| MW, pI | 45 kDa, 8.52 |
| Gene length, protein length | 1239 bp, 413 aa |
| Immediate neighbours | ywtE, capE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 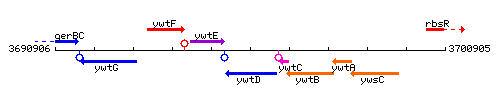
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed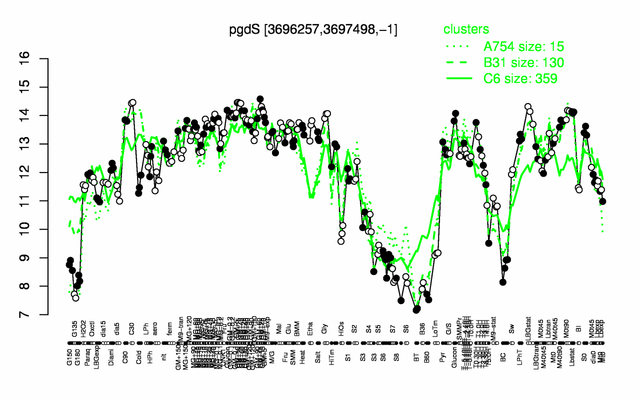
| |
Contents
Categories containing this gene/protein
capsule biosynthesis and degradation
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35860
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: nlpC/p60 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: secreted (according to Swiss-Prot), extracellular (signal peptide) PubMed
Database entries
- Structure:
- UniProt: P96740
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: pgdS PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed