Difference between revisions of "BesA"
| Line 22: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[dhbA]]'', ''[[yuiH]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[dhbA]]'', ''[[yuiH]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU32010 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU32010 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU32010 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU32010 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU32010 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU32010 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:yuiI_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:yuiI_context.gif]] | ||
Revision as of 11:10, 14 May 2013
- Description: trilactone hydrolase, catalyses ferri-bacillibactin hydrolysis leading to cytosolic iron release
| Gene name | besA |
| Synonyms | yuiI |
| Essential | no |
| Product | trilactone hydrolase |
| Function | iron acquisition |
| Gene expression levels in SubtiExpress: besA | |
| MW, pI | 35 kDa, 8.203 |
| Gene length, protein length | 942 bp, 314 aa |
| Immediate neighbours | dhbA, yuiH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 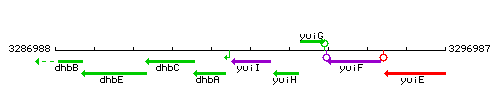
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
acquisition of iron, iron metabolism
This gene is a member of the following regulons
AbrB regulon, Fur regulon, Efp-dependent proteins
The gene
Basic information
- Locus tag: BSU32010
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O32102
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Rebecca J Abergel, Anna M Zawadzka, Trisha M Hoette, Kenneth N Raymond
Enzymatic hydrolysis of trilactone siderophores: where chiral recognition occurs in enterobactin and bacillibactin iron transport.
J Am Chem Soc: 2009, 131(35);12682-92
[PubMed:19673474]
[WorldCat.org]
[DOI]
(I p)
Marcus Miethke, Oliver Klotz, Uwe Linne, Jürgen J May, Carsten L Beckering, Mohamed A Marahiel
Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis.
Mol Microbiol: 2006, 61(6);1413-27
[PubMed:16889643]
[WorldCat.org]
[DOI]
(P p)
Noel Baichoo, Tao Wang, Rick Ye, John D Helmann
Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon.
Mol Microbiol: 2002, 45(6);1613-29
[PubMed:12354229]
[WorldCat.org]
[DOI]
(P p)