Difference between revisions of "TnrA"
(→References) |
|||
| Line 87: | Line 87: | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' feedback-inhibited [[GlnA]] prevents TnrA from DNA binding | + | * '''Effectors of protein activity:''' feedback-inhibited [[GlnA]] prevents [[TnrA]] from DNA binding |
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** [[TnrA]]-[[NrgB]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17001076 PubMed] | ** [[TnrA]]-[[NrgB]] [http://www.ncbi.nlm.nih.gov/sites/entrez/17001076 PubMed] | ||
| − | ** [[TnrA]]-[[GlnA]], this interaction results in loss of [[TnrA]] DNA-binding activity {{PubMed|11719184}} | + | ** [[TnrA]]-[[GlnA]], this interaction results in loss of [[TnrA]] DNA-binding activity as well as in inhibition of [[GlnA]] {{PubMed|23535029,11719184}} |
* '''[[Localization]]:''' membrane-associated via [[NrgA]]-[[NrgB]] under conditions of poor nitrogen supply {{PubMed|21435182}} | * '''[[Localization]]:''' membrane-associated via [[NrgA]]-[[NrgB]] under conditions of poor nitrogen supply {{PubMed|21435182}} | ||
| Line 157: | Line 157: | ||
<pubmed>11719184, 12139611, 17085574 19233925, 16885465, </pubmed> | <pubmed>11719184, 12139611, 17085574 19233925, 16885465, </pubmed> | ||
==Other original publications== | ==Other original publications== | ||
| − | <pubmed>12374841,15547269,9287005, 12950915,10671441,16547045,16547045 ,8799114, 15150225, 11029411,17001076,15547269, 2573733, 8636055, 16493705, 6141156 18667567 21435182</pubmed> | + | <pubmed>12374841,15547269,9287005, 12950915,10671441,16547045,16547045 ,8799114, 15150225, 11029411,17001076,15547269, 2573733, 8636055, 16493705, 6141156 18667567 21435182 23535029</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:27, 29 March 2013
- Description: transcriptional pleiotropic regulator invoved in global nitrogen regulation
| Gene name | tnrA |
| Synonyms | scgR |
| Essential | no |
| Product | transcription activator/ repressor |
| Function | regulation of nitrogen assimilation |
| Gene expression levels in SubtiExpress: tnrA | |
| Interactions involving this protein in SubtInteract: TnrA | |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Nucleotides (regulation), Ile, Leu, Val, Ammonium/ glutamate, Central C-metabolism, Cell wall, Coenzyme A, Phosphorelay, Alternative nitrogen sources | |
| MW, pI | 12 kDa, 10.235 |
| Gene length, protein length | 330 bp, 110 aa |
| Immediate neighbours | mgtE, ykzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 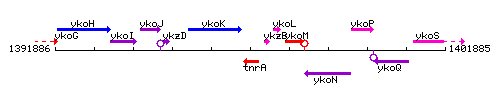
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed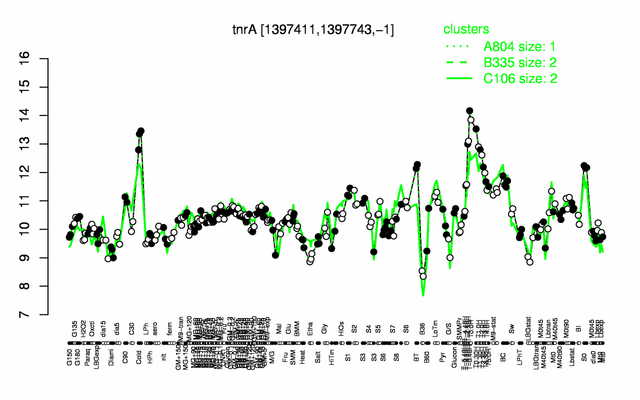
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, glutamate metabolism, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The TnrA regulon
The gene
Basic information
- Locus tag: BSU13310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- K(D) value for the binding site in the tnrA promoter region: 55 nM PubMed
- Domains:
- Modification:
- Cofactor(s):
- Localization: membrane-associated via NrgA-NrgB under conditions of poor nitrogen supply PubMed
Database entries
- Structure:
- UniProt: Q45666
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: tnrA (according to DBTBS)
- Regulation:
- Additional information:
Biological materials
- Mutant: GP252 (in frame deletion), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Karl Forchhammer lab
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
The TnrA regulon
Control of TnrA activity by the trigger enzyme GlnA
Other original publications