Difference between revisions of "FtsZ"
| Line 170: | Line 170: | ||
==Other original Publications== | ==Other original Publications== | ||
'''Additional publications:''' {{PubMed|22931116,22912848,21224850}} | '''Additional publications:''' {{PubMed|22931116,22912848,21224850}} | ||
| − | <pubmed> 15288790, 15317782,12180929, 9364910,10323866, 19212404,15942012, 12007411,16420366, 16159787,10747015, 16950129, 16796675,10322023, 9495766,9287012,1569582,10878122, 11395470,10449747,17662947, 12368265,18284588,8600030,18588879,7592498, 19136590 , 19429628, 19141479 19843223 16484179 20199598 20566861 20711458 20807205 20933427 15948963 12700262 22298780 22457634 22730127</pubmed> | + | <pubmed> 15288790, 15317782,12180929, 9364910,10323866, 19212404,15942012, 12007411,16420366, 16159787,10747015, 16950129, 16796675,10322023, 9495766,9287012,1569582,10878122, 11395470,10449747,17662947, 12368265,18284588,8600030,18588879,7592498, 19136590 , 19429628, 19141479 19843223 16484179 20199598 20566861 20711458 20807205 20933427 15948963 12700262 22298780 22457634 22730127 22984350 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:46, 21 September 2012
- Description: cell-division initiation protein (septum formation)
| Gene name | ftsZ |
| Synonyms | ts-1 |
| Essential | yes PubMed |
| Product | cell-division initiation protein (septum formation) |
| Function | formation of Z-ring |
| Gene expression levels in SubtiExpress: ftsZ | |
| Interactions involving this protein in SubtInteract: FtsZ | |
| MW, pI | 40 kDa, 4.814 |
| Gene length, protein length | 1146 bp, 382 aa |
| Immediate neighbours | ftsA, bpr |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 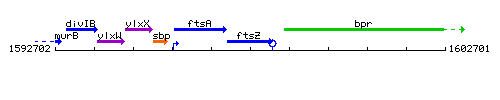
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed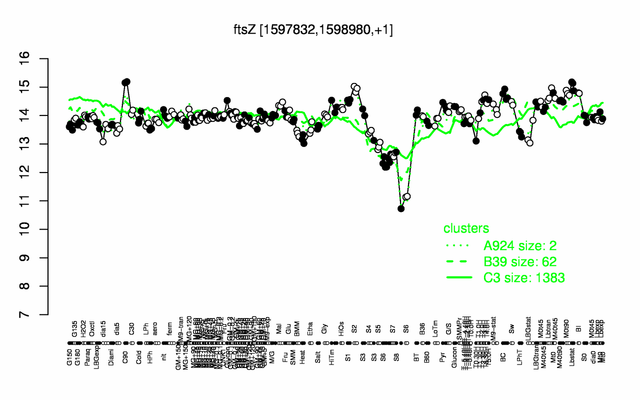
| |
Contents
Categories containing this gene/protein
cell division, essential genes, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15290
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ftsZ family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Z ring formation is inhibited upon binding of MciZ to FtsZ
- bundling of FtsZ protofilaments into strikingly long and regular tubular structures reminiscent of eukaryotic microtubules requires the prior formation of large ring polymers of SepF PubMed
- interaction with UgtP inhibits FtsZ filament formation PubMed
Database entries
- UniProt: P17865
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Jeff Errington lab
Labs working on this gene/protein
- Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
- Leendert Hamoen, CBCB, Newcastle University, UK
Your additional remarks
References
Reviews
Additional reviews: PubMed
An-Chun Chien, Norbert S Hill, Petra Anne Levin
Cell size control in bacteria.
Curr Biol: 2012, 22(9);R340-9
[PubMed:22575476]
[WorldCat.org]
[DOI]
(I p)
Christine Kaimer, Peter L Graumann
Players between the worlds: multifunctional DNA translocases.
Curr Opin Microbiol: 2011, 14(6);719-25
[PubMed:22047950]
[WorldCat.org]
[DOI]
(I p)
FtsZ as antibacterial drug target
Other original Publications
Additional publications: PubMed