Difference between revisions of "PMAD"
| Line 3: | Line 3: | ||
'''General comments''' | '''General comments''' | ||
| − | A [[plasmid]] for | + | This site is under construction |
| − | naturally nontransformable gram-positive bacteria. This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl- | + | |
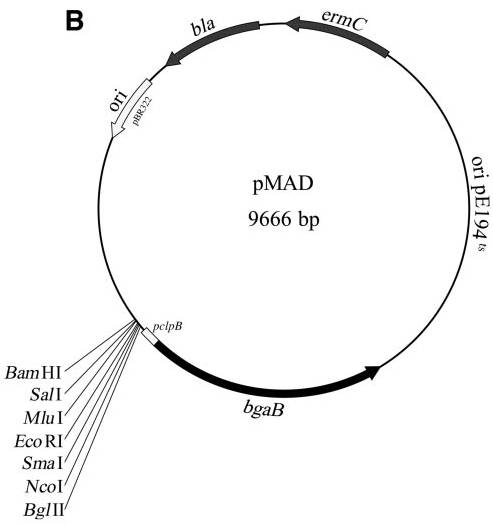
| − | beta-D-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the | + | A [[plasmid]] for generating gene inactivation mutants in naturally nontransformable gram-positive bacteria. |
| − | plasmid, facilitating clone identification during mutagenesis. The plasmid is can be used in Staphylococcus | + | This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the plasmid, facilitating clone identification during mutagenesis. |
| − | aureus, Listeria monocytogenes, and Bacillus spp. to efficiently construct mutants with or without an associated | + | The plasmid is can be used in Staphylococcus aureus, Listeria monocytogenes, and Bacillus spp. to efficiently construct mutants with or without an associated |
antibiotic resistance gene. | antibiotic resistance gene. | ||
* ''E.coli'': multicopy plasmid, select for ampicillin | * ''E.coli'': multicopy plasmid, select for ampicillin | ||
| − | * ''B. subtilis'': temperature sensitive ori, select for erythromycin/ lincomycin | + | * ''B. subtilis'': temperature-sensitive ori, select for erythromycin/ lincomycin |
'''Hints for the usage''' | '''Hints for the usage''' | ||
Revision as of 08:25, 28 August 2012
General comments
This site is under construction
A plasmid for generating gene inactivation mutants in naturally nontransformable gram-positive bacteria. This vector allows, on X-Gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) plates, a quick colorimetric blue-white discrimination of bacteria which have lost the plasmid, facilitating clone identification during mutagenesis. The plasmid is can be used in Staphylococcus aureus, Listeria monocytogenes, and Bacillus spp. to efficiently construct mutants with or without an associated antibiotic resistance gene.
- E.coli: multicopy plasmid, select for ampicillin
- B. subtilis: temperature-sensitive ori, select for erythromycin/ lincomycin
Hints for the usage
- for a markerless deletion of a gene amplify 1000 bp up- and downstream of the gene and clone them into the MCS of the plasmid
The reference