Difference between revisions of "YciC"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 1: | Line 1: | ||
| − | + | * '''Description:''' putative metallochaperone <br/><br/> | |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || zinc uptake | |style="background:#ABCDEF;" align="center"|'''Function''' || zinc uptake | ||
| − | |||
| − | |||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 4.399 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 45 kDa, 4.399 | ||
Revision as of 11:06, 7 August 2012
- Description: putative metallochaperone
| Gene name | yciC |
| Synonyms | |
| Essential | no |
| Product | putative metallochaperone |
| Function | zinc uptake |
| MW, pI | 45 kDa, 4.399 |
| Gene length, protein length | 1191 bp, 397 aa |
| Immediate neighbours | yciB, yckA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 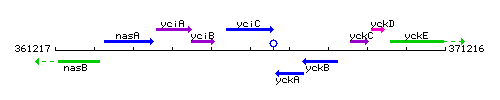
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed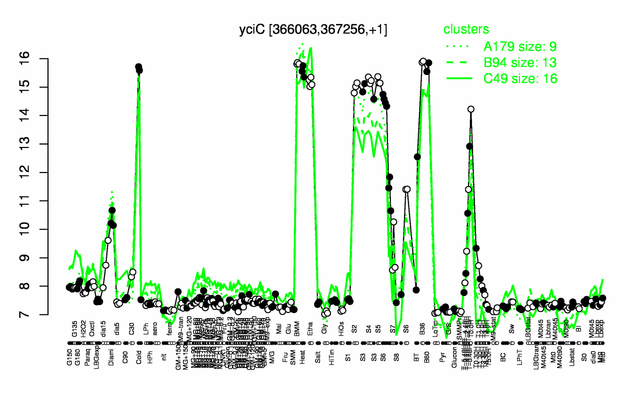
| |
Contents
Categories containing this gene/protein
trace metal homeostasis (Cu, Zn, Ni, Mn, Mo), membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03360
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: cobW C-terminal domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-58 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane associated PubMed
Database entries
- Structure:
- UniProt: P94400
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Scott E Gabriel, Faith Miyagi, Ahmed Gaballa, John D Helmann
Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur.
J Bacteriol: 2008, 190(10);3482-8
[PubMed:18344368]
[WorldCat.org]
[DOI]
(I p)
Ahmed Gaballa, Tao Wang, Rick W Ye, John D Helmann
Functional analysis of the Bacillus subtilis Zur regulon.
J Bacteriol: 2002, 184(23);6508-14
[PubMed:12426338]
[WorldCat.org]
[DOI]
(P p)
A Gaballa, J D Helmann
Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis.
J Bacteriol: 1998, 180(22);5815-21
[PubMed:9811636]
[WorldCat.org]
[DOI]
(P p)