Difference between revisions of "FloA"
(→Expression and regulation) |
(→Categories containing this gene/protein) |
||
| Line 40: | Line 40: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| + | {{SubtiWiki category|[[membrane dynamics]]}}, | ||
{{SubtiWiki category|[[cell envelope stress proteins (controlled by SigM, V, W, X, Y)]]}}, | {{SubtiWiki category|[[cell envelope stress proteins (controlled by SigM, V, W, X, Y)]]}}, | ||
{{SubtiWiki category|[[resistance against toxins/ antibiotics]]}}, | {{SubtiWiki category|[[resistance against toxins/ antibiotics]]}}, | ||
Revision as of 17:30, 6 July 2012
- Description: flottilin-like protein (in addition to FloT), resistence protein (against sublancin), accessory role in resistance to cefuroxime
| Gene name | floA |
| Synonyms | yqfA |
| Essential | no |
| Product | flottilin-like protein |
| Function | resistance against sublancin |
| MW, pI | 35 kDa, 4.917 |
| Gene length, protein length | 993 bp, 331 aa |
| Immediate neighbours | yqfB, yqeZ |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 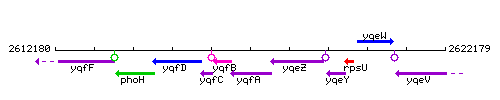
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
membrane dynamics, cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against toxins/ antibiotics, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25380
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: UPF0365 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54466
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Felix Dempwolff, Heiko M Möller, Peter L Graumann
Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins.
J Bacteriol: 2012, 194(17);4652-61
[PubMed:22753055]
[WorldCat.org]
[DOI]
(I p)
Daniel López, Roberto Kolter
Functional microdomains in bacterial membranes.
Genes Dev: 2010, 24(17);1893-902
[PubMed:20713508]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Bronwyn G Butcher, John D Helmann
Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli.
Mol Microbiol: 2006, 60(3);765-82
[PubMed:16629676]
[WorldCat.org]
[DOI]
(P p)
Min Cao, Phil A Kobel, Maud M Morshedi, Ming Fang Winston Wu, Chris Paddon, John D Helmann
Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches.
J Mol Biol: 2002, 316(3);443-57
[PubMed:11866510]
[WorldCat.org]
[DOI]
(P p)