Difference between revisions of "ScoA"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || lipid metabolism | |style="background:#ABCDEF;" align="center"|'''Function''' || lipid metabolism | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://cellpublisher.gobics.de/subtiexpress/ ''Subti''Express]''': [http://cellpublisher.gobics.de/subtiexpress/bsu/BSU38990 scoA] | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 25 kDa, 6.613 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 25 kDa, 6.613 | ||
Revision as of 17:26, 7 August 2012
- Description: probable succinyl CoA:3-oxoacid CoA-transferase (subunit A)
| Gene name | scoA |
| Synonyms | yxjD |
| Essential | no |
| Product | probable succinyl CoA:3-oxoacid CoA-transferase (subunit A) |
| Function | lipid metabolism |
| Gene expression levels in SubtiExpress: scoA | |
| MW, pI | 25 kDa, 6.613 |
| Gene length, protein length | 714 bp, 238 aa |
| Immediate neighbours | scoB, yxjC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 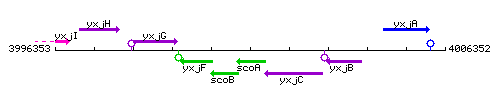
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed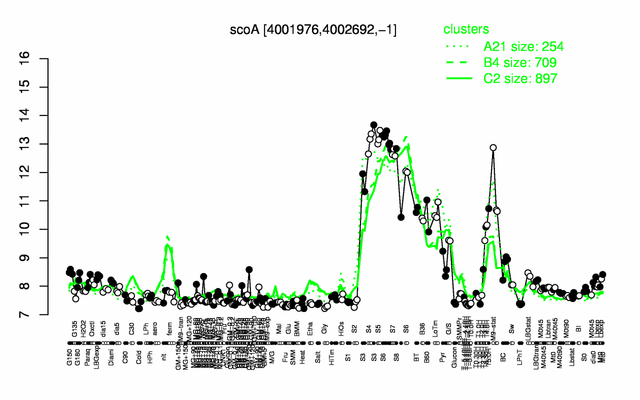
| |
Contents
Categories containing this gene/protein
utilization of lipids, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38990
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Succinyl-CoA + a 3-oxo acid = succinate + a 3-oxoacyl-CoA (according to Swiss-Prot)
- Protein family: 3-oxoacid CoA-transferase subunit A family (according to Swiss-Prot)
- Paralogous protein(s): YodS
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3CDK
- UniProt: P42315
- KEGG entry: [3]
- E.C. number: 2.8.3.5
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lucy Stols, Min Zhou, William H Eschenfeldt, Cynthia Sanville Millard, James Abdullah, Frank R Collart, Youngchang Kim, Mark I Donnelly
New vectors for co-expression of proteins: structure of Bacillus subtilis ScoAB obtained by high-throughput protocols.
Protein Expr Purif: 2007, 53(2);396-403
[PubMed:17363272]
[WorldCat.org]
[DOI]
(P p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Shane T Jensen, Erin M Conlon, Christiaan van Ooij, Jessica Silvaggi, José Eduardo González-Pastor, Masaya Fujita, Sigal Ben-Yehuda, Patrick Stragier, Jun S Liu, Richard Losick
The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis.
J Mol Biol: 2003, 327(5);945-72
[PubMed:12662922]
[WorldCat.org]
[DOI]
(P p)
Ken-Ichi Yoshida, Izumi Ishio, Eishi Nagakawa, Yoshiyuki Yamamoto, Mami Yamamoto, Yasutaro Fujita
Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome.
Microbiology (Reading): 2000, 146 ( Pt 3);573-579
[PubMed:10746760]
[WorldCat.org]
[DOI]
(P p)
Y Miwa, A Nakata, A Ogiwara, M Yamamoto, Y Fujita
Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis.
Nucleic Acids Res: 2000, 28(5);1206-10
[PubMed:10666464]
[WorldCat.org]
[DOI]
(I p)