Difference between revisions of "TnrA"
| Line 149: | Line 149: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 10231480 18086213 </pubmed> | + | <pubmed> 10231480 18086213 22625175</pubmed> |
==The [[TnrA regulon]]== | ==The [[TnrA regulon]]== | ||
<pubmed>12823818,</pubmed> | <pubmed>12823818,</pubmed> | ||
Revision as of 16:14, 28 May 2012
- Description: transcriptional pleiotropic regulator invoved in global nitrogen regulation
| Gene name | tnrA |
| Synonyms | scgR |
| Essential | no |
| Product | transcription activator/ repressor |
| Function | regulation of nitrogen assimilation |
| Interactions involving this protein in SubtInteract: TnrA | |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Nucleotides (regulation), Ile, Leu, Val, Ammonium/ glutamate, Central C-metabolism, Cell wall, Coenzyme A, Phosphorelay, Alternative nitrogen sources | |
| MW, pI | 12 kDa, 10.235 |
| Gene length, protein length | 330 bp, 110 aa |
| Immediate neighbours | mgtE, ykzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 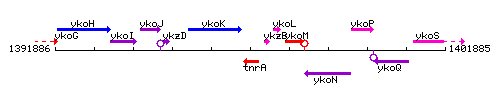
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed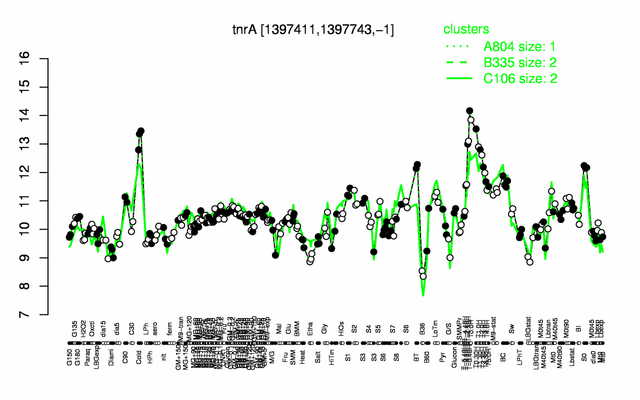
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The TnrA regulon
The gene
Basic information
- Locus tag: BSU13310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: feedback-inhibited GlnA prevents TnrA from DNA binding
- Localization: membrane-associated via NrgA-NrgB under conditions of poor nitrogen supply PubMed
Database entries
- Structure:
- UniProt: Q45666
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: tnrA (according to DBTBS)
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant: GP252 (in frame deletion), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Karl Forchhammer lab
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
The TnrA regulon
Control of TnrA activity by the trigger enzyme GlnA
Other original publications
Additional publications: PubMed
Airat Kayumov, Annette Heinrich, Margarita Sharipova, Olga Iljinskaya, Karl Forchhammer
Inactivation of the general transcription factor TnrA in Bacillus subtilis by proteolysis.
Microbiology (Reading): 2008, 154(Pt 8);2348-2355
[PubMed:18667567]
[WorldCat.org]
[DOI]
(P p)
Annette Heinrich, Kathrin Woyda, Katja Brauburger, Gregor Meiss, Christian Detsch, Jörg Stülke, Karl Forchhammer
Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis.
J Biol Chem: 2006, 281(46);34909-17
[PubMed:17001076]
[WorldCat.org]
[DOI]
(P p)
Jill M Zalieckas, Lewis V Wray, Susan H Fisher
Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA.
J Bacteriol: 2006, 188(7);2578-85
[PubMed:16547045]
[WorldCat.org]
[DOI]
(P p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Shigeo Tojo, Takenori Satomura, Kaori Morisaki, Ken-Ichi Yoshida, Kazutake Hirooka, Yasutaro Fujita
Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA.
J Bacteriol: 2004, 186(23);7971-9
[PubMed:15547269]
[WorldCat.org]
[DOI]
(P p)
Boris R Belitsky, Abraham L Sonenshein
Modulation of activity of Bacillus subtilis regulatory proteins GltC and TnrA by glutamate dehydrogenase.
J Bacteriol: 2004, 186(11);3399-407
[PubMed:15150225]
[WorldCat.org]
[DOI]
(P p)
Emmanuel Guedon, Charles M Moore, Qiang Que, Tao Wang, Rick W Ye, John D Helmann
The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons.
Mol Microbiol: 2003, 49(6);1477-91
[PubMed:12950915]
[WorldCat.org]
[DOI]
(P p)
Jaclyn L Brandenburg, Lewis V Wray, Lars Beier, Hanne Jarmer, Hans H Saxild, Susan H Fisher
Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter.
J Bacteriol: 2002, 184(21);6060-4
[PubMed:12374841]
[WorldCat.org]
[DOI]
(P p)
B R Belitsky, L V Wray, S H Fisher, D E Bohannon, A L Sonenshein
Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression.
J Bacteriol: 2000, 182(21);5939-47
[PubMed:11029411]
[WorldCat.org]
[DOI]
(P p)
D Robichon, M Arnaud, R Gardan, Z Pragai, M O'Reilly, G Rapoport, M Débarbouillé
Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-phoR global regulators.
J Bacteriol: 2000, 182(5);1226-31
[PubMed:10671441]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, S H Fisher
Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H.
J Bacteriol: 1997, 179(17);5494-501
[PubMed:9287005]
[WorldCat.org]
[DOI]
(P p)
L V Wray, A E Ferson, K Rohrer, S H Fisher
TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(17);8841-5
[PubMed:8799114]
[WorldCat.org]
[DOI]
(P p)
S W Brown, A L Sonenshein
Autogenous regulation of the Bacillus subtilis glnRA operon.
J Bacteriol: 1996, 178(8);2450-4
[PubMed:8636055]
[WorldCat.org]
[DOI]
(P p)
H J Schreier, S W Brown, K D Hirschi, J F Nomellini, A L Sonenshein
Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene.
J Mol Biol: 1989, 210(1);51-63
[PubMed:2573733]
[WorldCat.org]
[DOI]
(P p)
S H Fisher, A L Sonenshein
Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression.
J Bacteriol: 1984, 157(2);612-21
[PubMed:6141156]
[WorldCat.org]
[DOI]
(P p)