Difference between revisions of "Tig"
| Line 24: | Line 24: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:tig_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:tig_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=tig_2886315_2887589_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:tig_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/> | <br/><br/> | ||
Revision as of 10:20, 20 April 2012
- Description: trigger factor (prolyl isomerase)
| Gene name | tig |
| Synonyms | yzzH |
| Essential | no |
| Product | trigger factor (prolyl isomerase) |
| Function | protein folding |
| MW, pI | 47 kDa, 4.224 |
| Gene length, protein length | 1272 bp, 424 aa |
| Immediate neighbours | clpX, ysoA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 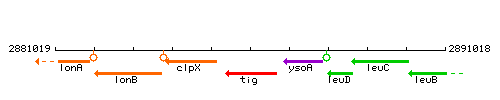
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed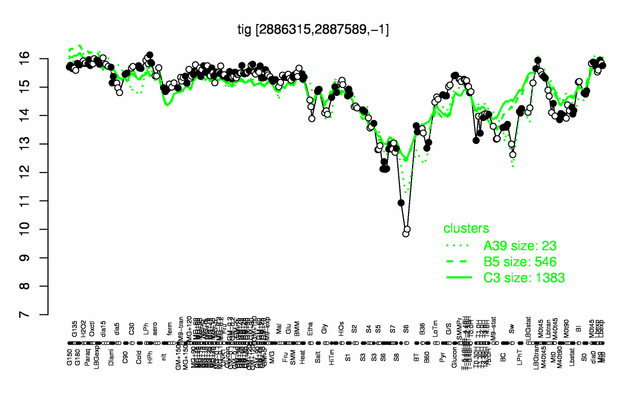
| |
Contents
Categories containing this gene/protein
chaperones/ protein folding, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU28230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Tig subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on ser/ thr/ tyr PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P80698
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Operon: tig PubMed
- Sigma factor:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Frieder Merz, Daniel Boehringer, Christiane Schaffitzel, Steffen Preissler, Anja Hoffmann, Timm Maier, Anna Rutkowska, Jasmin Lozza, Nenad Ban, Bernd Bukau, Elke Deuerling
Molecular mechanism and structure of Trigger Factor bound to the translating ribosome.
EMBO J: 2008, 27(11);1622-32
[PubMed:18497744]
[WorldCat.org]
[DOI]
(I p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Dindo Y Reyes, Hirofumi Yoshikawa
DnaK chaperone machine and trigger factor are only partially required for normal growth of Bacillus subtilis.
Biosci Biotechnol Biochem: 2002, 66(7);1583-6
[PubMed:12224648]
[WorldCat.org]
[DOI]
(P p)
S F Göthel, C Scholz, F X Schmid, M A Marahiel
Cyclophilin and trigger factor from Bacillus subtilis catalyze in vitro protein folding and are necessary for viability under starvation conditions.
Biochemistry: 1998, 37(38);13392-9
[PubMed:9748346]
[WorldCat.org]
[DOI]
(P p)
S F Göthel, R Schmid, A Wipat, N M Carter, P T Emmerson, C R Harwood, M A Marahiel
An internal FK506-binding domain is the catalytic core of the prolyl isomerase activity associated with the Bacillus subtilis trigger factor.
Eur J Biochem: 1997, 244(1);59-65
[PubMed:9063446]
[WorldCat.org]
[DOI]
(P p)
A Wipat, N Carter, S C Brignell, B J Guy, K Piper, J Sanders, P T Emmerson, C R Harwood
The dnaB-pheA (256 degrees-240 degrees) region of the Bacillus subtilis chromosome containing genes responsible for stress responses, the utilization of plant cell walls and primary metabolism.
Microbiology (Reading): 1996, 142 ( Pt 11);3067-78
[PubMed:8969504]
[WorldCat.org]
[DOI]
(P p)